Ali Navid (14-ERD-091)
Abstract
Engineering of microbial consortia is a new frontier in synthetic biology. By programming the conduct and performance of select microbial communities, we can force these organisms to coordinate their efforts to achieve a specific goal such as production of compounds of interest like biofuels or drugs. Engineering multicellular communities to achieve a specific goal requires system-level understanding of the workings and capabilities of each organism in the community and their interactions. Computational models are usually used to conduct such system-level analyses. Unfortunately, available modeling tools and methods are limited to examining only one objective of the system, while analysis of multicellular communities requires developmental models that conduct multiple-objective flux analysis of the system. Our primary goal with this research project is to develop an algorithm for generation of multiple-objective flux analysis models for analysis of interactions among cells in multicellular communities. We will use this method to optimize biofuel production in a synthetic co-culture of mutant strains of Clostridium phytofermentans, an anaerobic, rod-shaped bacterium capable of producing ethanol and hydrogen gas.
The success of this research project will result in (1) development of a computational tool for generation of genome-scale multiple-objective flux analysis models that can be run using the Laboratory's high-performance computers, (2) added insight into central carbon metabolism of biofuel-producing organisms, (3) establishment of a novel metabolically engineered consortium of multiple strains of C. phytofermentans that have been optimized for peak production of ethanol, and (4) a system-level analysis of multicellular communities that greatly benefit microbial consortia engineering efforts. Developing a tool for automatic generation of genome-scale multiple-objective flux analysis models of metabolism will be of great utility for systems biology studies of multicellular systems. Coupling this progress to the Laboratory's extensive computational capabilities will place us at the forefront of examining and engineering complex multicellular systems.
Mission Relevance
This project aligns well with LLNL's core competency in bioscience and bioengineering, as well as the strategic focus area of chemical and biological security. Results of our development of an algorithm for automatic generation of models that would account for different metabolic objectives of diverse members of a microbial community can be used for the development of new countermeasures against biosecurity threats and examination of interactions between immune system and pathogens.
FY15 Accomplishments and Results
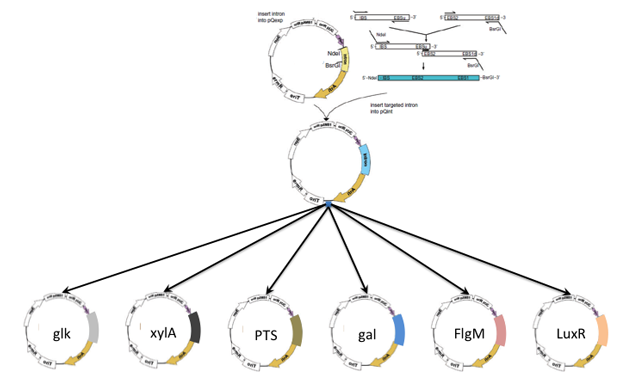
In FY15 we (1) employed metabolite measurements of C. phytofermentans batch experiments grown in the presence of select hexose or pentose sugars to constrain a genome-scale computer model for conducting a system-level analysis of metabolism; (2) identified a set of target genes that will be inactivated to generate sugar substrate-specific mutants, as a result of our systems-level analyses; (3) used the CRISPR/Cas9 technology (clustered regularly interspaced short palindromic repeats) of gene manipulation to target the genes responsible for consumption of glucose and xylose; (4) verified the phenotype of sugar-specific strains during growth experiments, and compared growth curves; and (5) began developing the code for multiple-objective flux analyses of our synthetic microbial consortium.