Bradley Hart (15-FS-010)
Abstract
The Forensic Science Center at Lawrence Livermore plays an important role in providing technical support to the nation's response to terrorist use of weapons of mass destruction, and is one of two U.S. laboratories internationally certified for identifying chemical warfare agents. In addition, it has performed pivotal biological and chemical forensics analyses in many high-profile criminal investigations. The center has developed novel protein-based human identification methods for use in forensic analysis, and has focused on the use of hair shaft proteins (primarily keratins and other structural proteins) as the source of genetically variant peptides used in identification (peptides are basically chains of amino acids that are smaller than protein chains.) We proposed to examine the feasibility of using tissues other than hair to yield proteins that are useful for this analysis. Specifically, we explored the feasibility of extracting proteins from teeth and bones, analysis of these proteins, and bioinformatics of the resulting data. We processed modern and archaeological teeth (both male and female) and analyzed the resulting peptide samples using a Thermo Q-Exactive Plus Mass Spectrometer. The protein population in each sample included sex-specific peptides from the protein amelogenin, which allowed for unambiguous sex determination of both modern and archeological teeth. When bone samples were analyzed using the same methods, they were found to contain genetically variant peptides and many common proteins observed in teeth. Based on the results of this study, we can conclude that the tooth and bone proteome can be good candidates for the identification of these peptides, which in turn can be used to calculate measures of identity and biogeographic background. We also found that the tooth proteome can be used to accurately determine sex using both modern and archeological teeth.
Background and Research Objectives
Livermore's Forensic Science Center has established a research program to identify genetic features in proteins called single amino-acid polymorphisms, and to use this information to calculate measures of individual identity and biogeographic background.1,2 To date, method development has focused on hair shafts as a source of forensically relevant protein material. We have identified, characterized, and validated over 60 genetic markers in hair protein. These genetic peptide markers can be used to infer the status of single-nucleotide polymorphism alleles in a subject’s genome. Using this methodology, we have obtained individual powers of discrimination of up to 1 in 5.5 million. This data set has also demonstrated the power of the approach to indicate biogeographic origin. The collection of identified genetic markers in these samples was 2,400 times more likely to occur in the European sample population than the African. These measures of identity combined with likelihood ratio values have a variety of forensic applications. They allow for exclusion of an individual and complement other forensic measures of identity, such as mitochondrial haplotype analysis (a set of DNA variations, or polymorphisms, that tend to be inherited together) and short tandem repeat-profiles.
While hair is particularly robust, often only teeth and bone are available to the investigator. Because of environmental exposure, DNA may have degraded below the limit of detection. In this case, we hypothesized that genetically quantifiable information can still be retained in the protein population of teeth and bone. Analyzing these proteins may yield forensically useful identifying information that is statistically valid.
Teeth highly express a protein that has two isoforms, one that resides on the X-chromosome and one on the Y-chromosome.3,4 Theoretically, unique peptides from one or both of these isoforms could be used to determine the sex of an individual. This gene family has long been used to sex biological material using DNA-based methods.3 We hypothesized that sex determination of human remains using teeth could occur using protein-based methods. This possibility has not been systematically tested and only very limited reporting of proteomic identifications from such samples are available in the literature.5,6
The possibility of protein-based sex determination is important because the traditional means of determining the sex of skeletons—visual and biometric analysis of anatomical features—has major limitations. Sex determination of sub-adult, juvenile, and incomplete skeletons cannot be performed accurately.7 In teeth we have the possibility of using sex-specific peptides to identify sex of previously unknown skeletons. This technique provides an additional layer of interpretation to archaeological remains and allows for comparison with other features such as osteopathology, biometrics, and cultural context.
Scientific Approach and Accomplishments
We hypothesized that teeth and bone could be used as a source of genetically informative peptides, and that teeth could be used to determine sex of individuals. To test these hypotheses, the following tasks were developed:
Task 1: Determine the feasibility of obtaining genetically informative peptides from modern human bone, archaeological human teeth, and modern human teeth.
Task 2: Determine feasibility of improving datasets with the Thermo Q-Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer.
Task 3: Establish the feasibility of applying novel bioinformatics protocols for analysis of genetically variant peptides using proteomic software packages.
Results
Small blocks of human bone and enamel tissue (20 mg) were dissected and subjected to a series of chemical and physical processes to facilitate analysis. The resulting sample was prepared by proteolysis overnight in the presence of trypsin. The resulting peptide solution was then applied to the Thermo Q-Exactive Plus Mass Spectrometer for proteomic analysis.
Bone and Teeth Proteomes
The bone and teeth proteomes showed considerable similarity. Two high-quality bone proteomes were obtained in the study, with 186 proteins and 1,400 different peptides identified in a modern sample using The Global Proteome Machine open-source bioinformatics tool (peptide-sequence matching software) for a modern sample, and 153 proteins and 1,062 different peptides identified in an archaeological sample (250–450 years old). A disproportionate amount of protein signal was centered on two gene products, collagen type 1 α1 and type 1 α2. Other collagens also were highly represented in the remaining protein population (Table 1). The modern bone, consistent with its collection from freshly obtained autopsy material, also contained a significant number of blood and bone marrow proteins. Similar numbers were obtained for the teeth proteomes: 195 and 176 proteins for male and female teeth, corresponding to 1,073 and 1,478 different peptides, respectively. Both modern and archaeological teeth protein populations were, like bone, dominated by collagens, particularly type 1 α1 and α2, and extracellular matrix proteins. Proteins specific for enamel biogenesis, maintenance, and regulation, such as ameloblastin, enamelysin, and amelogenin X and Y were also identified, as were some salivary proteins, such as lysozyme.
Table 1. The most abundant proteins in bone for both female and male teeth proteomes. Blood and marrow proteins are indicated in red and enamel-specific proteins are indicated in bold black. Note the presence of AMELY in the male tooth proteome (#21). Novel genes include a descriptor (gene: description).
| Female Tooth | Male Tooth | Bone | |
| 1 | COL1A1: collagen, type I, α1 | COL1A1 | COL1A1 |
| 2 | COL1A2: collagen, type I, α2 | COL1A2 | COL1A2 |
| 3 | COL5A2: collagen, type V, α2 | COL5A2 | COL5A2 |
| 4 | ALB: albumin | COL5A1 | COL7A1: p, collagen, type VII, α1 |
| 5 | COL3A1: collagen, type III, α1 | AHSG | HBA2 |
| 6 | COL5A1: collagen, type V, α1 | ALB | HIST1H1C: p, histone cluster 1, H1c |
| 7 | COL4A5: collagen, type IV, α5 | AMBN | COL5A1 |
| 8 | COL6A6: collagen, type VI, α6 | COL3A1 | HBB: hemoglobin, beta |
| 9 | AHSG: alpha-2-HS-glycoprotein | AMELX | HIST1H1D: histone cluster 1, H1d |
| 10 | AMBN: Ameloblastin | COL11A2 | HIST1H1A: histone cluster 1, H1a |
| 11 | COL4A4: collagen, type IV, α4 | COL4A4 | VTN |
| 12 | COL9A3: collagen, type IX, α3 | BGN | BGLAP: Polyamine-modulated factor 1 |
| 13 | AMELX: amelogenin, X-linked | ENAM | HIST1H2BL: histone cluster 1, H2bl |
| 14 | COL11A2: collagen, type XI, α2 | COL9A3 | COL3A1 |
| 15 | BGN: biglycan | F2 | COL14A1: collagen, type XIV, α1 |
| 16 | HBA2: hemoglobin, alpha 2 | KNG1: kininogen 1 | HIST1H1E: histone cluster 1, H1e |
| 17 | F2: thrombin | COL17A1: collagen, type XVII, alpha 1 | ACTB: actin, beta |
| 18 | SKIV2L2: superkiller viralicidic activity 2-like 2 | SERPINA1: alpha-1 antiproteinase | BGN: biglycan |
| 19 | IgG heavy chain | VTN: vitronectin | ACTG1: actin gamma 1 |
| 20 | VIM: vimentin | MMP20 | HIST1H2BA: histone cluster 1, H2ba |
| 21 | VTN: vitronectin | AMELY: amelogenin, Y-linked | AHSG |
Genetically Variant Peptides
Candidate genetically variant peptides were identified in each tissue proteome. Peak lists from each bone and teeth trypsin-digest sample were analyzed using The Global Proteome Machine that identifies candidate single amino-acid variants. Peptides associated with two single amino-acid polymorphisms were identified in the peptide populations in the respective teeth and bone data sets (Table 2). The most abundant genetically variant peptides contain a polymorphism corresponding to position in collagen type 1 α2 (P549A, rs42524). Both alleles of this highly common polymorphism (minor genotypic frequency = 42%) were detected in both proteomes. A rare genetically variant peptide in the abundant blood protein ASHG (R317C, rs35457250 with minor genotypic frequency = 1.5%) was also detected in one of the tooth proteomes. Further expansion of the suite of genetically variant peptides detected in each proteome will require steps at both the sample-processing and data-acquisition stages to maximize coverage of each proteome for both tissues. Other bone proteomic studies, using two-dimensional-gels and multiple mass spectrometry applications, have identified over 2,400 different protein families, indicating that there remains significant room for identification and development of additional genetically variant peptide signatures.8
Table 2. Candidate genetically variant peptides from teeth and bone. The genes (GN) and corresponding single-nucleotide polymorphism (rs#) are listed, along with the resulting single amino-acid polymorphism (SAP). The detection of each peptide in a bone (B) or tooth (T) proteome is indicated. The distribution of each peptide, known as genotypic frequency (GF), in sample European (EUR) or African (AFR) populations is also listed. The SAP is indicated in red with minor alleles represented in lower case. * Indicates the presence of a SAP preceding the observed peptide sequence.
| Tissue |
Genotypic Frequency |
||||||
| GN | rs# | Peptide Sequence | SAP | B | T | EUR | AFR |
| COL1A2 | rs42524 | GEQGPPGPPGFQGLPGPSGPAGEVGKPGER | P549A | X | X | 0.936 | 0.967 |
| GEQGPaGPPGFQGLPGPSGPAGEVGKPGER | X | X | 0.439 | 0.201 | |||
| GAPGPDGNNGAQGPPGPQGVQGGKGEQGP PGPPGFQGLPGPSGPAGEVGKPGER |
X | X | 0.936 | 0.967 | |||
| GAPGPDGNNGAQGPPGPQGVQGGKGEQGP aGPPGFQGLPGPSGPAGEVGKPGER |
X | X | 0.439 | 0.201 | |||
| AHSG | rs35457250 | *HTFMGVVSLGSPSGEVSHPR | R317C | 0 | X | 1.000 | 1.000 |
| AHYDLcHTFMGVVSLGSPSGEVSHPR | 0 | X | 0.016 | 0.001 | |||
Using Sex-Specific Peptides to Determine Sex from Teeth
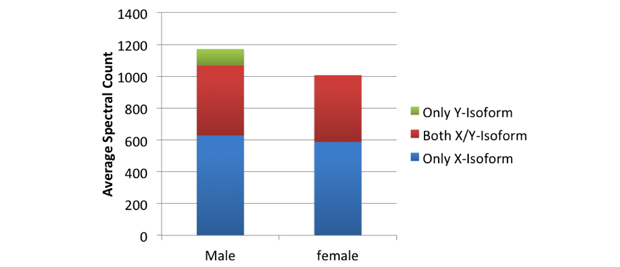
Tooth enamel is produced, maintained, and regenerated using enamel-specific proteins and enzymes. One of these proteins, amelogenin, has specific isoforms that occur on each of the two sex-determining chromosomes.4 Unambiguous identification of specific X- or Y-isoforms therefore allows us to determine a karyotype for these individuals. Initial analysis of trypsin digests of enamel in a male subject identified peptides that were specific for both the X- and Y-isoform of amelogenin (Figure 1), as well as those that were common in both isoforms.5,6 Cumulative information from analysis of several samples allowed us to develop a comprehensive inclusion list of amelogenin peptides. This list was then used to specifically target amelogenin peptides for fragmentation during mass spectrometry analysis, dramatically increasing the sensitivity of the method. Based on the presence of peptides specific for the Y-isoform of amelogenin, we were able to sex 7 male teeth using this technique with 100% specificity and 100% sensitivity. These peptides were completely absent in the female teeth sample (Figure 1). Given that the false negative rate was 0% for male teeth in this cohort, absence of the Y-isoform was indicative of female sex (with a sensitivity and specificity of 100%).
Reported tooth and dentin proteomes typically do not include amelogenin.5–9 This is most likely because of the unique biology of the molecule, which makes observation by classical proteomic methods highly challenging. For example, a study in which the protein was successfully identified exploited acid etching over a short time frame.5,6 In this case, only a single peptide was observed and only in a fraction of analyzed samples. Given the many processing interventions and steps utilized in that particular report, we hypothesized that acid etching, combined with a simpler sample-processing protocol, with minimal opportunities for peptide loss, would provide the greatest probability of success. As described here, this hypothesis proved correct and we were able to identify a comprehensive set of amelogenin peptides.
Impact on Mission
Our effort addresses gaps in forensic science in support of the Laboratory's strategic focus area of cyber security, space, and intelligence, particularly with respect to providing novel forensic methods with applications in counterproliferation, counterterrorism, and domestic security. The research also is directly relevant to the core competency in bioscience and bioengineering. The feasibility of forensic methods and applications resulting from this effort may enable the development of an entirely new tool set for use in intelligence, law enforcement, homeland security, and healthcare.
Conclusion
We have shown previously that detection of genetically variant peptides can be a basis for forensic analysis of hair proteins. Here we establish that tooth and bone proteomes, which persist in the environment even longer than hair, can also be a source of these peptides, with two candidates identified. Sex-specific peptides were also detected in the tooth proteome with 100% specificity and 100% sensitivity. This has the potential to revolutionize forensic and physical anthropology, because there are currently no methods to accurately sex juvenile or disrupted or degraded skeletons. We made full use of the recently acquired Thermo Q-Exactive Plus Mass Spectrometer located in the Forensic Science Center. This current state-of-the-art instrument allowed for efficient exploitation of both DNA informed or “top-down” and unbiased or “bottom-up” data-acquisition strategies. The bottom-up acquisition was highly effective in detection of novel peptides and identifying members of the protein population in each sample type. The resulting inclusion list was highly sensitive at the top-down targeted detection of sex-specific peptides. The use of multiple approaches extended to bioinformatic analysis of the data sets. The Global Proteome Machine was efficient at detection of candidate genetically variant peptides, and the PEAKS proteomics mass spectrometry software was efficient at peptide spectral matching of detected amelogenin peptides.
References
- Parker, G. J., Methods for conducting genetic analysis using protein polymorphisms, U.S. Patent #US8877455 B2 (2014).
- Parker, G. J., et al. Demonstration of protein-based human identification using the hair shaft proteome. (2015).
- Tsai, T. C., et al., “Identification of sex-specific polymorphic sequences in the goat amelogenin gene for embryo sexing.” J. Anim. Sci. 89(8), 2407 (2011).
- Salido, E. C., et al., “The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes.” Am. J. Hum. Genet. 50(2), 303 (1992).
- Porto, I. M., et al., “Recovery and identification of mature enamel proteins in ancient teeth.” Eur. J. Oral Sci. 119 (suppl. 1), 83 (2011).
- Porto, I. M., et al., “Techniques for the recovery of small amounts of mature enamel proteins.” J. Arch. Sci. 38, 3596 (2011).
- Saunders, S. R., “Juvenile skeletons and growth-related studies.” Biological anthropology of the human skeleton, second edition. John Wiley & Sons, Inc., Hoboken, NJ (2008).
- Jiang, X., et al., “Method development of efficient protein extraction in bone tissue for proteome analysis”, J. Proteome Res. 6(6), 2287 (2007).
- Jagr, M., et al., “Comprehensive proteomic analysis of human dentin.” Eur. J. Oral Sci. 120(4), 259 (2012).