Maxim Shusteff (14-LW-077)
Abstract
Recent decades have seen enormous leaps forward in DNA sequencing, bioinformatics, and viral genetics. Despite these advances, viral infectious diseases continue to present major public health threats worldwide, as well as to U.S. national security. A major reason that effective therapies and countermeasures remain extraordinarily challenging to develop is that viruses are dynamic systems, while most tools for studying them are static. Studying viruses in vitro significantly distorts their infection patterns, evolutionary parameters, and replication dynamics. Where animal models exist, experimental flexibility is significantly more limited than in vitro. A bridging capability is needed that can combine the control and flexibility of in vitro environments with more realistic in vivo system dynamics. Our main goal with this research project is to apply microfluidic acoustic filtering to build a viral culture platform that more realistically mimics the dynamic equilibrium of in vivo infection, compared with standard in vitro culture methods. The proposed system will use acoustic separation to clear free virus particles and re-circulate infected and uninfected host cells back into the culture, establishing the first quasi-steady-state viral culture. In collaboration with world-class virologists, we will demonstrate the utility of the system for investigating different host–virus systems.
Successful completion of this project will result in a bridging technology, filling the capability gap between static viral culture (simple and inaccurate) and animal models or clinical studies of human patients (complex and costly). We will create a microfluidic device for development of an equilibrium culture system. To accomplish the influx of fresh cells required simultaneously with clearance of viral particles, we will couple the microfluidic filter to a commercial mammalian cell-culture bioreactor that will deliver uninfected lymphocytes to the culture to replenish cells killed by viral infection. The new viral culturing platform, together with the associated mathematical tools, will allow entirely new measurements of evolutionary selection parameters, host-cell response, drug effectiveness, and the emergence of drug resistance in viral infections. Most broadly, the success of this work will enable a radically new paradigm in studying infectious disease, and enhance the development of effective and safe therapies.
Mission Relevance
This work is relevant to LLNL's biosecurity mission to rapidly mitigate evolving and unknown biological threats, of which diagnostic platforms and understanding of host–pathogen interactions are key enabling capabilities. These capabilities support the Laboratory's core competency in bioscience and bioengineering. We anticipate establishing new technologies in the realms of pathogen detection and characterization and host–pathogen interactions that will impact development of countermeasures to biological threats. Federal agencies such as the Defense Advanced Research Agency and National Institutes of Health have likewise identified the development of new platforms for drug development as a key mission area.
FY15 Accomplishments and Results
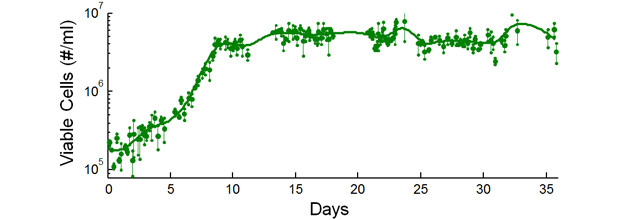
In FY15 we (1) optimized and validated our acoustic focusing device for its high-performance, cell–virus separation capabilities at some of the highest throughput ever reported for acoustic microfluidic separators; (2) designed and configured an automated, recirculating mammalian cell-culture system with extensive real-time monitoring and feedback; and (3) confirmed that the system was able to maintain cell growth over week-long timescales, which represents the longest continuous operation of microfluidic separation devices reported to date (see figure).
Publications and Presentations
- Bora, M., et al. “Efficient coupling of acoustic modes in microfluidic channel devices.” Lab Chip 15, 3192 (2015). LLNL-JRNL-664577. http://dx.doi.org/10.1039/c5lc00343a
- Fong, E. J., et al., “A microfluidic platform for precision small-volume sample processing and its use to size separate biological particles with an acoustic microdevice.” J. Vis. Exp. doi:10.3791/53051 (2015). LLNL-JRNL-665235.
- Jung, S. Y., et al., “Spatial tuning of acoustofluidic pressure nodes by altering net sonic velocity enables high-throughput, efficient cell sorting.” Lab Chip 15, 1000 (2015). LLNL-JRNL-665514. http://dx.doi.org/10.1039/c4lc01342e