Jeffrey Drocco (16-FS-023)
Project Description
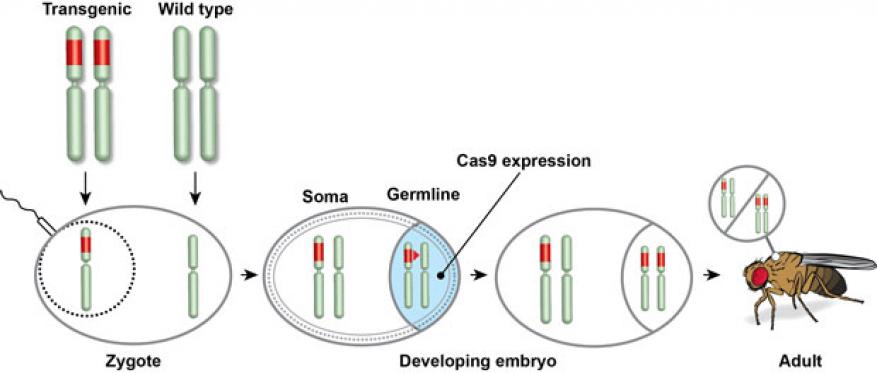
Recently, there has been renewed interest in the use of molecular genetics techniques to create gene drives—that is, means of introducing novel genes into reproducing populations that are subsequently propagated to high frequency (shown in figure). This genetic tool could, for example, be used in wild populations to help control malaria. Using an infectious deoxyribonucleic acid (DNA) construct that spreads through mosquito chromosomes and progeny alike, researchers could conceivably create a population of biting insects that could be resistant to the malaria-causing parasite. The recent discovery of CRISPR-Cas9 genome editing prompted the demonstration earlier this year of a highly efficient gene drive, or mutagenic chain reaction, in the fruit fly. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR-associated (Cas) genes enable organisms to respond to and eliminate invading genetic material. The Cas9 protein has been heavily utilized as a genome engineering tool to induce site-directed double strand breaks in DNA, which can lead to gene inactivation or introduction of genes derived from a different organism. This technology shows great promise for controlling crop pests and disease vectors, but scientists have raised concerns about the possibility of either intentional or unintentional ecological damage from the use of these techniques. We plan to integrate experimental results with computational simulation to explore the feasibility of a prototype modeling capability that will enable understanding of the spread of an introduced gene drive. This may include models that account for the durability of an alternative gene from mutagenic chain reaction, variations in the susceptibility of individuals within the population to mutagenic chain reaction mutagenesis, and propagation through spatially constrained populations.
Gene drives are a potentially powerful innovation with wide-ranging economic and environmental implications. We expect to identify the key parameters underpinning the susceptibility of natural populations to modification via a gene drive mechanism, as well as assess the feasibility of measuring these parameters to construct a model capable of estimating the likelihood of success or failure of an introduced mutagenic chain reaction in such populations. We intend to advance current modeling capabilities beyond the Unckless analytical model, which provides a basic understanding of the propagation of a mutagenic chain reaction but does not account for the possibility of resistance to the drive or for nonrandom mating arising from spatial constraints that may cause the reaction to cease propagating in a local area. An improved modeling capability would allow for better assessment of the risks and benefits of future uses of this new technology.
Mission Relevance
This project is aligned with Livermore's core competency in chemical and biological security, and builds on its scientific strengths in the areas of detailed consequence modeling using high-performance computing. The mutagenic chain reaction is a new technology with potentially significant ecological effects arising from the manipulation of natural populations. In addition, we believe the outcome of this feasibility study will be relevant to biosecurity or environmental protection challenges.
FY16 Accomplishments and Results
In FY16 we (1) computationally characterized the deviations from normal behavior of the susceptible, infected, and removed epidemic model when a gene drive mechanism creates the possibility of a mutation of the targeted species; (2) collaborated with experts from Cornell University in Ithaca, New York, and New York University in New York City in both the theoretical and experimental aspects of genetic drive technology; and (3) began modeling the propagation of a mutagenic chain reaction.