Daniel Faissol (16-FS-041)
Project Description
Agent-based models are a class of computational models that simulate the actions of autonomous agents (both individual or collective entities) to observe their effects on the system as a whole. Widely used in such fields as biology, social science including economics, and technology, agent-based models have demonstrated value in representing the complex behaviors that emerge from systems composed of agents following a simple set of rules. These models offer a promising approach for testing hypotheses of potential mechanisms in biological systems that contain stochasticity (a system lacking predictable order) and spatial heterogeneity, such as the immune system or cancerous tumors. Any network-based phenomenon can potentially be represented by way of these models. Agent-based models could advance Livermore’s security missions in biodefense, cybersecurity, and counterterrorism. However, users currently perform model calibration manually and qualitatively, severely limiting their use. To begin addressing this issue, we will explore the feasibility of multiple approaches for automatically calibrating these models at scale with synthetic data. We plan to study the feasibility of combining novel approaches developed in disparate disciplines to resolve the problem of parameter calibration of agent-based models. The study will be a critical first step to learning mechanisms in biological systems using agent-based models, machine learning, and stochastic optimization.
Successfully calibrating agent-based model parameters automatically (i.e., learning values that develop parameters for model rules) is a first step to learning mechanisms in biological systems with agent-based models (i.e., learning rules that govern the models). Therefore, this feasibility study will provide two related but distinct benefits. First, we will make the first-ever attempt at using the novel techniques specified below to automatically calibrate agent-based models to increase their scalability and predictive power. Second, we will gain a deeper understanding of the challenges that will be faced in future research, along with recommendations for addressing them. We will explore a Bayesian analysis system of statistical inference based on interpreting probability to model calibration, specifically, approximate Bayesian computation. While experiments have previously been done on this approach within the context of agent-based models, it is not clear how well the approach scales to handle large problems and complex data. We will examine the prospect of using this approach at scale for eventual use in the broader goal of learning mechanisms of biological systems. We will also explore the use of gradient-based and gradient-free optimization, implementing such novel approaches as simultaneous-perturbation stochastic approximation with compressive sensing, and parallel gradient-free optimization to learn the scaling limitations of these approaches and how they can be addressed in future work.
Mission Relevance
The research most directly develops the Laboratory’s core competency of bioscience and bioengineering through cutting-edge techniques in predictive simulation, and understanding and simulating complex biological systems. Our research into automatic calibration of computational agent-based models supports the core competency in high-performance computing, simulation, and data science. Additionally, this proposal supports Livermore’s mission in biological security. Enabling the development of truly predictive models in biology would allow the Laboratory to better develop countermeasures against various threats such as emerging pathogens and synthetic biological agents.
FY16 Accomplishments and Results
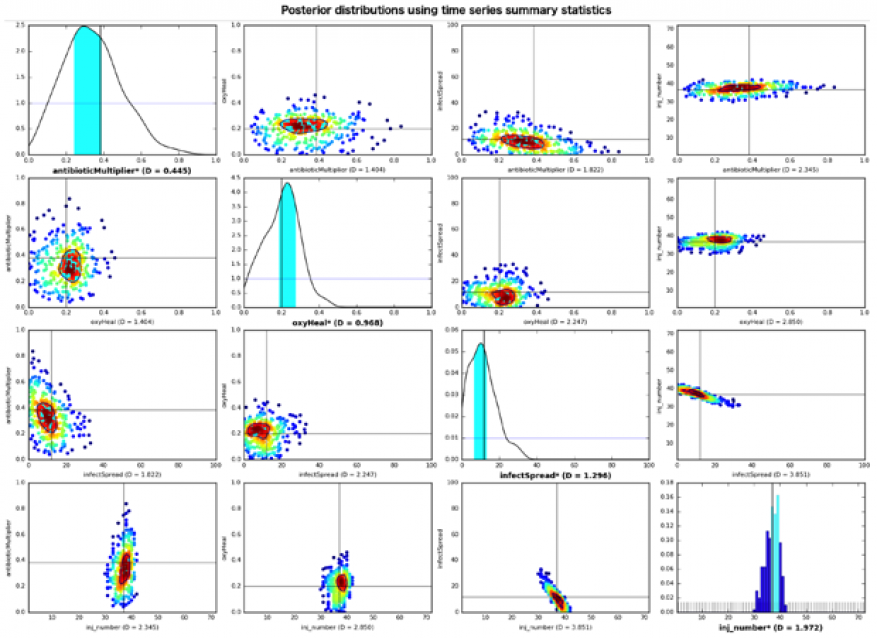
In FY16 we (1) implemented our University of Chicago collaborator’s agent-based simulation code on Livermore Computing machines and parallelized it to perform large-scale experiments; (2) implemented the approximate Bayesian computation algorithm on the same machines to calibrate the simulation using synthetic data, and ran experiments on both our collaborator’s simulation, as well as other open-source simulations; (3) demonstrated utility of the Bayesian approach by recovering parameter values used to generate simulation output with a high degree of accuracy for most of the parameter values with several different distance functions (see figure); and (4) implemented the simultaneous-perturbation stochastic approximation algorithm (method for optimizing systems with multiple unknown parameters), and began running experiments with the algorithm. The figure shows sample results from implementing the Approximate Bayesian Computation Rejection Algorithm to calibrate parameter values of an agent-based model of the innate immune response using synthetic temporal data. The results show that the parameter values were successfully calibrated within a reasonable error tolerance when using an L2-norm summary statistic for each variable and the standardized Euclidean distance metric.