Bradley Hart
Overview
Human hair contains minimal intact nuclear deoxyribonucleic acid (DNA) for human identification in forensic and archaeological applications. In contrast, proteins offer a pathway to exploit hair evidence for human identification, owing to their persistence, abundance, and derivation from DNA. Individualizing single nucleotide polymorphisms are often conserved as single amino acid polymorphisms in genetically variant peptides. Detection of genetically variant peptide markers in the hair proteome via high-resolution tandem mass spectrometry permits inference of single nucleotide polymorphisms with known statistical probabilities. The research conducted in this project has established an entirely new approach to human identification. The primary goals of this effort were to develop the foundational science to enable high-fidelity proteomic analysis of hair samples. To accomplish this, it was necessary to establish methods and approaches spanning a variety of technical disciplines including biochemistry, high-resolution mass spectrometry, computational proteomic and genomic analysis, and statistical methods. Ultimately this project resulted in published methods that will play a significant role in addressing serious and systemic issues with forensic science, namely an over-reliance on subjective methodology, by providing a statistically valid approach to evidence analysis that does not rely on DNA.
Background and Research Objectives
Use of human hair in forensics has contributed to investigations aimed at associating individuals with scenes and objects connected with a crime (Maxfield and Babbie 2014). Current forensic techniques applied to hair characterization include microscopic analysis and mitochondrial DNA (mtDNA) sequencing (Moeller, Fey, and Sachs 1993). Studies intended to understand the reliability of microscopic analysis reveal fundamental issues underlying the approach (Wennig 2000, Robertson 2002, Smith and Goodman 1995). For example, in 2002 an FBI study published a landmark report in forensic science that systematically and comprehensively analyzed a large collection of previous casework to measure the frequency of false-positive associations in microscopic comparisons (Houck and Budowle 2002). "Its conclusion was of enormous importance to forensic science, to police, to courts and to juries: When hair examiners conclude in casework that two hair samples are microscopically indistinguishable, the hairs often (1 in 9 times) come from different sources," (President’s Council of Advisors on Science 2016, Maxfield and Babbie 2014). This report, among others, details significant shortfalls stemming from the reliance of microscopic hair analysis on subjective systems of correlation, such as expert witness opinion (Wendler et al. 2010, Saks and Koehler 2005, Rowe 2001, Murphy 2007, Moriarty and Saks 2005). However, mtDNA is an objective alternative that can provide statistically relevant measures of human identity (Houck and Budowle 2002, Melton et al. 2005). The direct inheritance of mtDNA from the maternal genetic line (e.g. mother to child) limits the discriminatory power of this methodology (Melton 2016, Keastle et al. 2006). Additional forensic techniques that utilize hair evidence can increase the potential information gained during investigations.
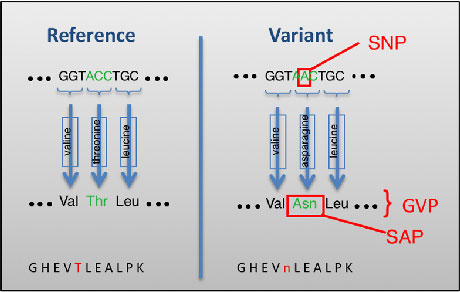
The research conducted under this project drove the development of a new forensic technique that exploits genetically variant peptides (GVPs) found in human hair and offers an alternative method that is both statistically relevant and objective (Parker et al. 2016b). As illustrated in the figure below, this approach probes genetic variations in humans by associating mutations found in hair proteins (single amino acid polymorphisms or "SAPs") to their DNA counterparts (single nucleotide polymorphisms or "SNPs") in protein-encoding regions (Parker et al. 2016b, Wu et al. 2017). The initial demonstration of this method characterized GVPs in 10 mg of human hair and implemented a “post-validation” approach (Parker et al. 2016b, Wu et al. 2017). The "post-validation"” technique refers to identification of potential GVP identity markers through proteomic analysis alone, followed by confirmation using genetic sequencing.
Additionally, a "pre-validation" approach, which involves the initial genetic sequencing of the whole exome (the part of the genome containing all the protein-coding genes) of a subject’s DNA, was used to later inform individualized SNP and SAP protein databases. Protein databases created from exome sequences were then programmed into proteomic software to identify potential GVPs that are genetically relevant in tandem mass spectrometry (MSMS) data collected on specific hair protein extractions. Consequently, this strategy has a built-in genetic validation component because only DNA-encoded mutations are identified. By gathering all possible protein mutations associated with a person’s DNA, the "pre-validation" approach greatly expands the opportunity to detect as many GVPs as possible. In combination, results from this two-tiered approach provided greater numbers of uniquely identified proteins and peptides than previously published. Additionally, more genetically relevant GVPs were identified. This study established that GVPs found in hair protein, as well as protein from other tissues, such as bone and teeth, can be a source of human identity markers. Additionally, this effort resulted in a deep understanding of the statistical basis for determining identity based on proteomic markers. We developed and published standardized protocols for sample processing and data analysis to allow implementation of this approach by the forensic science community.
Impact on Mission
Through this project, we established an entirely new approach to human identification. This research paves the way for addressing serious and systemic issues currently within the field of forensic science related to over-reliance on subjective methodology by providing a statistically valid approach to evidence analysis that does not rely on DNA. While DNA analysis is and will remain the gold standard for human identification, there are specific scenarios and issues, described earlier in this report, for which DNA analysis is challenged or unusable. Proteomic analysis of human tissues is a natural complement to DNA profiling in many ways.
Additionally, this project paved an entirely new technical path for the Forensic Science Center at Lawrence Livermore National Laboratory, driving the development of proteomic analysis tools, capabilities, infrastructure, and staff. Significant follow-on funding has been secured to further develop the science and applications associated with this approach.
Conclusion
The primary goal of this research was to discover completely new technical approaches to forensic identification of humans that are not reliant on subjective methodology, but rather build on a foundation of scientifically verifiable and statistically validated analytical methodology. The need for such science has been noted by the National Research Council as well as the President’s Council of Advisors on Science and Technology. We have shown that through the use of proteomic analysis, human identification can be conducted on multiple human tissue types with very high statistical confidence. Importantly, we have also shown that the sample sizes and types required for this analysis are consistent with realistic forensic scenarios.
References
Houck, M. M. and B. Budowle. 2002. "Correlation of Microscopic and Mitochondrial DNA Hair Comparisons." Journal of Forensic Science 47 (5):1–4.
Keastle, F. A., et al. 2006. "Database Limitations on the Evidentiary Value of Forensic Mitochondrial DNA Evidence. Am. Crim. L. Rev. 43:53.
Maxfield, M. G. and E. R. Babbie. 2014. Research Methods for Criminal Justice and Criminology. Nelson Education.
Melton, T. 2016. "Mitochondrial DNA: Profiling." Wiley Encyclopedia of Forensic Science.
Melton, T., et al. 2005. "Forensic Mitochondrial DNA Analysis of 691 Casework Hairs." Journal of Forensic Science 50 (1):JFS2004230-8.
Moeller, M. R., et al. 1993. "Hair Analysis as Evidence in Forensic Cases." Forensic Science International, 63, 43–53.
Moriarty, J. C. and M. J. Saks. 2005. "Forensic Science: Grand Goals, Tragic Flaws, and Judicial Gatekeeping." Judges J. 44:16.
Murphy, E. 2007. "The New Forensics: Criminal Justice, False Certainty, and the Second Generation of Scientific Evidence." California Law Review.
Parker, G. J., et al. 2016. "Demonstration of Protein-Based Human Identification Using the Hair Shaft Proteome." PloS One 11 (9):e0160653.
President's Council of Advisors on Science and Technology. 2016. Report to the President, Forensic Science in Criminal Courts: Ensuring Scientific Validity of Feature-comparison Methods. Executive Office of the President of the United States, President's Council of Advisors on Science and Technology.
Robertson, J. R. 2002. Forensic Examination of Hair. CRC Press.
Rowe, W. F. 2001. "The Current Status of Microscopical Hair Comparisons." The Scientific World.
Saks, M. J. et al. 2005. "The Coming Paradigm Shift in Forensic Identification Science." Science.
Smith, C. A. and P. D. Goodman. 1995. "Forensic Hair Comparison Analysis: Nineteenth Century Science or Twentieth Century Snake Oil." Colum. Hum. Rts. L. Rev. 27:227.
Wendler, C., et al. 2010. "The Path Forward: The Future of Graduate Education in the United States." Report from the Commission on the Future of Graduate Education in the United States. Princeton.
Wennig, R. 2000. "Potential Problems with the Interpretation of Hair Analysis Results." Forensic Science International 107 (1–3):5–12.
Wu, Pei-Wen, et al. 2017. "Proteomic Analysis of Hair Shafts From Monozygotic Twins: Expression Profiles And Genetically Variant Peptides." PROTEOMICS 17 (13–14):1600462. doi: 10.1002/pmic.201600462.
Publications and Presentations
Chu, F., et al. 2017. "Optimization of Single Hair Proteomics for Human Identification." Poster presentation, SciX 2017: The Federation of Analytical Chemistry and Spectroscopy Societies Conference, Reno, NV, 8–13 Oct. 2017. LLNL-POST-739124.
Chu, F., et al. 2018. "Assessment of Chemical Variation in the Hair Proteome from Different Body Locations Using Nano-LC-MS/MS for Human Identification." Oral presentation, 69th Annual Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, Orlando, FL, 26 Feb–1 March 2018. LLNL-PRES-746026.
Chu, F., et al. 2018. "Proteomic Variation Along the Length of Scalp Hair for Protein-Based Human Identification." Oral presentation, 66th American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics Annual Meeting, San Diego, CA, 3–7 June 2018. LLNL-PRES-751672.
Chu, F., et al. 2018. "Comparison of Protein and mtDNA Profiles for Protein-Based Human Identification." Poster presentation, Gordon Research Conference on Forensic Analysis of Human DNA, Newry, ME, June 2018. LLNL-POST-751843.
Hart, B. R., et al. 2017. "Methods and Systems to Perform Genetically Variant Protein Analysis, and Related Marker Genetic Protein Variations and Databases" U.S. Provisional Pat. Ser. No. 62/555,001, filed Sept. 6, 2017.
Kranes, S., et al. 2017. "Simultaneous DNA and Protein Extraction Using Trypsin." Forensic Science International: Genetics Supplement Series 6:e203-e204. doi: 10.1016/j.fsigss.2017.09.081. LLNL-ABS-739237.
Mason, K. E. et al. 2016. "Forensic Science of Genetically Variant Peptides." Oral presentation, Forensic Science in Defense and Security Symposium, Shrivenham, UK, 14–15 Sept. 2016. LLNL-PRES-680304.
Mason, K. E. et al. 2017. "Sex Determination: A Novel Protein-Based Sex Assignment Technique Using Human Tooth Enamel and Mass Spectrometry." Oral presentation, 69th Annual American Association of Forensic Science Conference, New Orleans, LA, 13–18 Feb. 2017. LLNL-ABS-699130.
Mason, K. E. et al, 2017. "Overview of Proteomic Approaches to Forensics at LLNL." Oral presentation. SciX 2017: The Federation of Analytical Chemistry and Spectroscopy Societies Conference, Reno, NV, 8–13 Oct. 2017. LLNL-PRES-739790.