Maxim Shusteff (15-LW-029)
Abstract
The use of large-scale computer simulations to study human physiology, especially for understanding disease states, is increasingly becoming a viable approach for the selection of therapeutic options and improving patient outcomes. However, the validity of such computational models (i.e., how well they represent real physical conditions) must be explored and demonstrated before they mature into useful predictive tools. This project undertook to develop and validate a massively-parallel fluid-dynamics simulation code for the purpose of modeling the movement of cancer cells in human blood vessels for predicting cancer metastasis. To collect accurate measurements within a physical model to which the computational models can be compared, we used three-dimensional (3D) printing to produce flow structures containing physiological-scale blood vessels, in which we experimentally measured fluid- and particle-flow parameters. Using vessel geometries representing the opposite ends of the vascular size spectrum, we investigated flow in the human aorta (approximately 1-cm vessel diameters) and in capillary networks (approximately 5–10 µm). The aorta was chosen as a representative large vessel for creating a metric to quantify the probability of metastatic cells to anchor in specific locations. Within capillaries, we explored the physics of normal and cancer cells squeezing through network constrictions, relating to similar considerations of cells arresting in specific locations. Each vascular-size scale presented different computational and in vitro experimental challenges, but we obtained promising results at each size scale. Using the combined capabilities of powerful computational models and advanced manufacturing technologies to solve difficult medical problems points toward a promising new direction for the future of medicine.
Background and Research Objectives
The role of computational fluid dynamics is becoming increasingly important in many fields, one of which is modeling circulatory diseases. Advances in high-performance computing (HPC) and multiphysics algorithms have made it possible to model fluid flow through a variety of complex geometries with unprecedented fidelity. Such models are quickly becoming more relevant as tools for understanding and predicting the progress of diseases, and eventually influencing therapeutic decisions. To attain these capabilities, the computational models must be validated (i.e., shown to be accurate within an acceptable range) for specific disease states.
Progress in additive manufacturing (AM) has made it possible to quickly and flexibly produce physical models of complex 3D computer-designed structures that include physiological geometries derived from patient-specific imaging data. The aim of this work was to combine Lawrence Livermore National Laboratory’s expertise in AM and HPC to create a new tightly-coupled computational validation pipeline. By conducting experiments using 3D-printed geometries generated from the same input geometries used in the fluid-dynamics simulations, our goal was to directly compare experimental results to theoretical projections. High-resolution simulations of explicitly-modeled cells in the human circulatory system were conducted on the Laboratory's Blue Gene/Q supercomputer.
In the initial phase of the project, we developed novel methods to study particle flow and cellular movement in large blood vessels with centimeter-scale diameters. Work during the third year of the project focused on extending our approaches to vessels at the capillary scale (approximate 5–10 µm diameter). Ultimately, we were able to demonstrate both computational model results and fabrication of vascular geometries at both macroscopic and microscopic scales, which yielded promising initial comparisons of in vitro and in silico data.
Scientific Approach and Accomplishments
For computational fluid-dynamics modeling in silico, we relied on HARVEY, a scalable code for producing massively parallel supercomputer simulations, recently developed by Professor Amanda Randles, a key project collaborator, currently at Duke University. HARVEY (named after the 17th-century surgeon, William Harvey, who first described the circulatory system) can model blood flow through macroscopic portions of the human circulatory system at micron-scale resolutions (Randles et al. 2013). For instance, HARVEY was used to demonstrate the first 3D simulation of flow in the full human arterial network down to the millimeter scale (Randles et al. 2015).
Large-Vessel Geometries
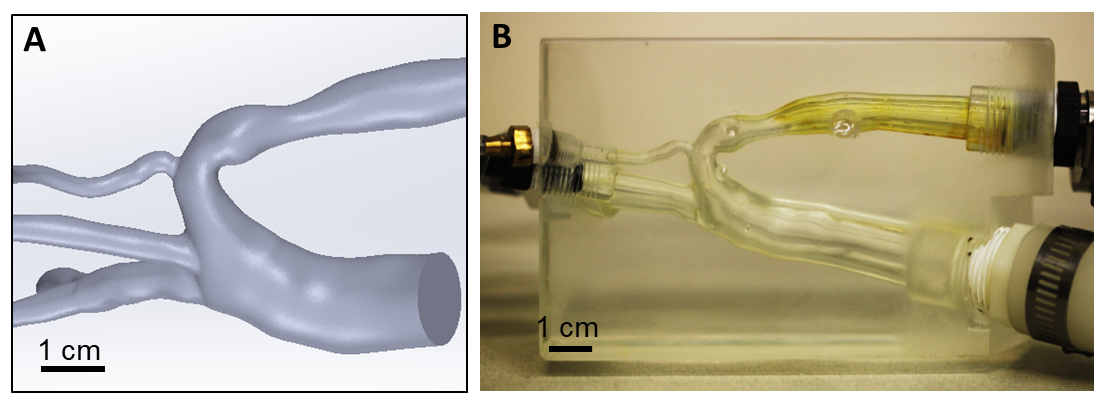
To carry out in vitro experiments, we fabricated optically clear vessels through which fluid flow could be visualized and which enabled us to use particle-tracking techniques. For large vessels, we chose a geometry that included a coarctation (i.e., a constriction) of the human aorta (Figure 1A). In collaboration with Professor David Frakes of Arizona State University, we produced a 3D model (Gounley et al. 2016) in clear urethane (Figure 1B) that was connected to tubing and a pump within a flow loop, allowing imaging of fluid loaded with fluorescent particles.
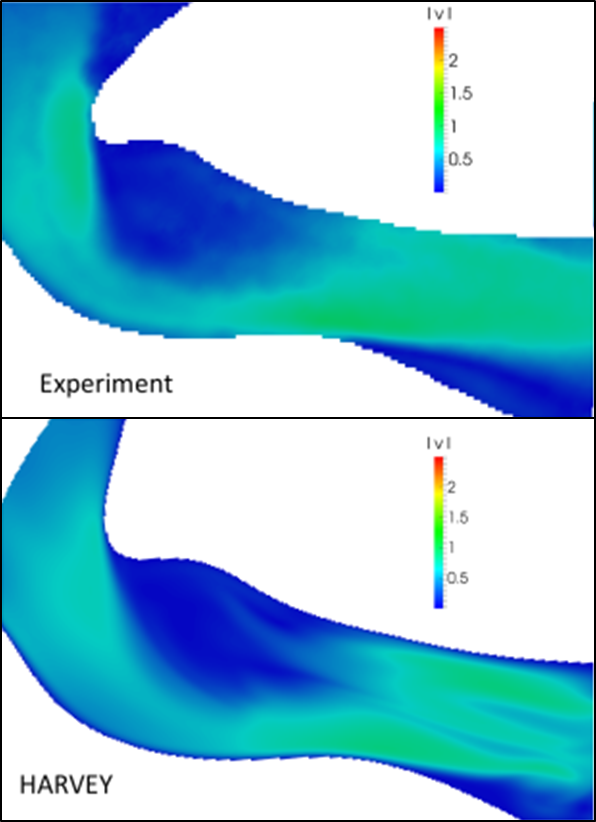
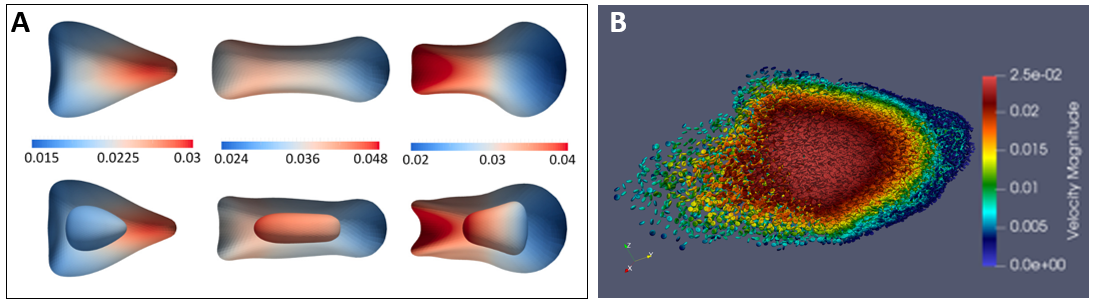
We designed a method based on light-sheet illumination to track cell trajectories and velocities in 3D throughout large blood vessel models (Laurence et al. 2017). We experimentally recapitulated both steady and pulsatile fluid flow in the large artery, validating the computed flow fields (Figure 2). A model of deformable cells was added to the supercomputer simulation (to represent both blood and cancer cells), for which validation is ongoing.
Figure 2. Experimental (top) and computational (bottom) flow field comparison. The flow velocity field was experimentally measured in m/s by particle image velocimetry (PIV) in the urethane model shown in Figure 1. Model validation continues.
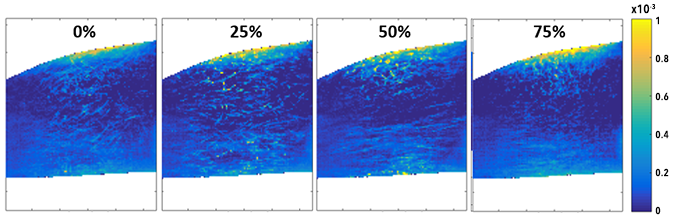
Figure 3 shows a plot of the experimentally-generated occupational probability function (OPF, which represents the relative probability of tracer particles being found at various locations in the geometry) near the aortic coarctation, for which the comparison to computationally-predicted locations of cancer cell dwell-time is ongoing.
Microscale Capillary Vessels
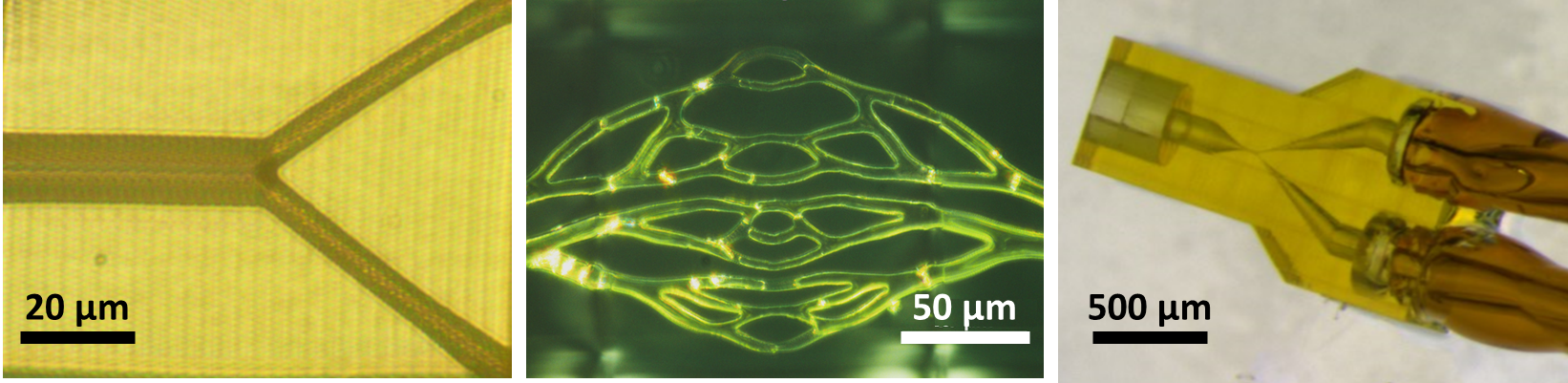
The majority of commercial high-end 3D printers have a minimum single-layer thickness of 10–20 µm, making them unsuitable for creating capillary-vessel models, for which the internal diameter is on the same size scale. To overcome this problem, we took advantage of Livermore’s recent acquisition of a 3D direct laser writing (DLW) system capable of 200 nm resolution and suitable for printing microvasculature. Although the DLW method is optimal for writing lattice-type and filament- or beam-based structures, rather than fluid channels (which are voids within solid blocks), we were able to fabricate simple and more complex capillary-vessel geometries on this system (Figure 4). These parts included integrated port structures suitable for interfacing the approximately 10-µm scale of individual vessels to 0.1–1mm scale external tubing (Figure 4, right).
The reliability of creating fully-open channels, which can be imaged for particle tracking, remains a work in progress. Likewise, the process of making fluidic connections still needs to be improved in order to routinely incorporate these structures into an instrumented flow loop.
When complete, this experimental data will help validate computational simulations in the capillary-vessel regime. Modeling the cancer cell as a compound capsule, we investigated the influence of the inner capsule (representing the cell nucleus) as the cell transits a narrow capillary-like constriction (Gounley et al. 2017) as shown in Figure 5A. Future comparisons with the experimental data generated here will improve model validation in this regime, particularly in capillary beds. The physiological behavior in complex capillary beds is of particular interest for understanding cancer cell arrest, as most current simulations and experiments only consider idealized or simplified geometries. In preparation for this experimental comparison, the computational model has been elongated to more easily see the large number of red blood cells in whole blood (Figure 5B).
Impact on Mission
This project equally supports three of Lawrence Livermore National Laboratory's core competencies: HPC, AM, and bioscience/bioengineering. The computer-based simulation and HPC techniques and capabilities developed and implemented during this project are now being used to support the Laboratory’s biosecurity and bioengineering program areas. The positive impact that our work has had on the growth of these program areas is evident, as these capabilities have now been incorporated into ongoing bio-printing research projects, as well as new work building an instrumented ex vivo tumor platform. More broadly, this work has helped to position the Laboratory to participate in larger external collaborative efforts that bring computational, AM, and in vitro capabilities together to study critical problems in medicine. One example is the Accelerating Therapeutics for Opportunities in Medicine initiative co-sponsored by the Laboratory, the National Cancer Institute, and GlaxoSmithKline.
Conclusion
This project has opened up initial efforts in a new and extremely fertile research space, in which cutting-edge computer modeling, data processing, and fabrication capabilities are combined to tackle the most pressing challenges in biomedicine. The current status and opportunities of these approaches has recently been described in a prominent review article (Randles et al. 2017).
The work begun on this project is continuing toward refining a suite of optimized AM-fabrication approaches spanning the micro-to-macro scales and including the meso-scale, as well as toward corresponding developments of blood-flow and deformable cell models at multiple simulation scales. At the conclusion of the project, our collaborative team, in partnership with a group of clinicians, submitted an R01 proposal to the National Cancer Institutes (PA-16-160), aiming to use these tools to improve treatment for pediatric congenital heart abnormalities. Livermore’s strong collaborative partnership with Professor Randles and Duke University enables the close coordination between computation and experiment that is required to obtain new kinds of data at previously unattainable fidelity. We expect to continue pursuing other opportunities for collaboration with the National Institutes for Health in the near term.
References
Gounley, J., et al. 2016. “Does the Degree of Coarctation of the Aorta Influence Wall Shear Stress Focal Heterogeneity?” 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), Orlando, Florida, August 2016. doi: 10.1109/EMBC.2016.7591465.
——— 2017. “Numerical Simulation of a Compound Capsule in a Constricted Microchannel.” Procedia Computer Science 108: 175–84. doi: 10.1016/j.procs.2017.05.209. LLNL-PRES-733050.
Laurence, T. A., et al. 2017. “Using Stroboscopic Flow Imaging to Validate Large-Scale Computational Fluid Dynamics Simulations.” Proceedings of SPIE 10076. doi: 10.1117/12.2253319. LLNL-PRPC-736453.
Randles, A., 2013. “Performance Analysis of the Lattice Boltzmann Model Beyond Navier-Stokes.” Proceedings of the 27th International Symposium on Parallel & Distributed Processing (IPDPS): 1063–74. IEEE. doi: 10.1109/IPDPS.2013.109.
Randles, A., et al. 2015. “Massively Parallel Models of the Human Circulatory System.” Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. doi: 10.1145/2807591.2807676. LLNL-CONF-670030.
——— 2017. “Computational Fluid Dynamics and Additive Manufacturing to Diagnose and Treat Cardiovascular Disease.” Trends in Biotechnology 35 (11): 1049–61. doi: 10.1016/j.tibtech.2017.08.008.
Publications and Presentations
Gounley, J., et al. 2017. “Numerical Simulation of a Compound Capsule in a Constricted Microchannel.” Procedia Computer Science 108: 175–84. doi: 10.1016/j.procs.2017.05.209. LLNL-PRES-733050.
Laurence, T. A., et al. 2017. “Using Stroboscopic Flow Imaging to Validate Large-Scale Computational Fluid Dynamics Simulations.” Proceedings Volume 10076, High-Speed Biomedical Imaging and Spectroscopy: Toward Big Data Instrumentation and Management II (100761E). doi: 10.1117/12.2253319. LLNL-PRPC-736453.
Randles, A., et al. 2015. “Massively Parallel Models of the Human Circulatory System.” Supercomputing 2015: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. doi: 10.1145/2807591.2807676. LLNL-CONF-670030.