Rhona Stuart (16-LW-030)
Abstract
Growing biofuel crops on marginal lands is our best option for sustainable biofuel production that does not interfere with food production, however nitrogen fertilizer requirements are a major barrier to sustainable practices. A grand challenge in agriculture is to partner agricultural plants with microbes that can fix nitrogen, thus eliminating the need for nitrogen fertilization, but manipulating known N-fixing symbionts to partner with new plants has proved challenging. In this project, we characterized a less studied class of N-fixing symbionts, from the genus Nostoc, in order to provide new potential target organisms for manipulation. Few plant-microbe symbioses are fully characterized at the molecular level, mostly due to the lack of approaches and tools, so this research also focused on applying novel approaches that would be applicable to other symbioses. During this project, we were able to (1) demonstrate that this symbiosis is a new type of plant-microbe N symbiosis, (2) identify resource fluxes between partners required for mutualism, (3) develop the tools to verify novel fluxes, and (4) conduct experiments to measure fluxes between elemental partners (Fe, S, N, and C). We established a molecular interaction model that provides metabolite targets to perturb symbiont response to the native host, as well as expression targets to measure response. Our findings represent a key departure from known diazotroph-host interactions, and imply that our symbiont may be more "independent" from its host.
Background and Research Objectives
Federal legislation has mandated that approximately 25 percent of liquid transportation fuel must come from non-grain biofuels by 2022 (Congress 2007). While this could be achieved by high-productivity crops cultivated on fertile land, it would be at the expense of the nation’s food supply (Valentine et al. 2012). An alternative that has attracted interest and garnered support is growing cellulosic biofuels sustainably on marginal lands (Tilman et al. 2006, Gelfand et al. 2013, DOE 2014). A major barrier to this alternative is the necessity for nitrogen fertilization of these crops. Nitrogen fertilization causes numerous problems that include (1) expense that cuts into the low profit margin of biofuel production; (2) increased N2O flux, which can negate greenhouse gas benefits of these fuels (Gelfand et al. 2013, Tilman et al. 2006); and (3) widespread damage to coastal ecosystems and fisheries through nitrogen runoff, resulting in eutrophication and aquatic “dead zones” (Diaz and Rosenberg 2008).
Biological nitrogen fixation (N-fixation), which delivers biologically regulated N inputs, provides a potential solution to this problem (Andrews et al. 2003, Cocking 2012). Diazotrophs are microorganisms that convert atmospheric N2 gas to NH3 and often live in close association with plant hosts. However, N-fixation is harmful to diazotrophs, requiring energy, metal cofactors (Fe and Mo), and protection from oxygen. To offset these costs, diazotrophs engage in complex symbiotic interactions with surrounding organisms, including the plant host. The best-studied diazotroph symbionts, rhizobia (legume root-nodule bacteria), are highly specific to their host, due largely to highly evolved plant immune responses (Berrabah et al. 2015), which has hindered attempts to transfer these symbionts into other plants (Stokstad 2016), despite substantial agricultural investment. More promising are recently described epiphytic diazotrophs, where the symbiont resides on the plant surface, as opposed to within a specialized compartment. However, these epiphytic symbioses are largely uncharacterized, especially at the molecular level, limiting our ability to study and manipulate them.
We chose to study a particular epiphyte, a cyanobacterium that associates with feather mosses, a member of the genus Nostoc (hereafter referred to simply as “Nostoc”). In boreal forests, these Nostoc species form symbiotic associations with feather mosses and provide the largest input of fixed-N into these systems (up to 50 percent of total N input; DeLuca et al. 2002 and 2008). While the feather moss–cyanobacteria association has been well studied at the ecological level, it is not well characterized at the molecular level. Our project sought to characterize epiphytic diazotrophic symbioses applying a combination of (1) comparative physiological experiments and gene expression analyses; and (2) stable-isotope tracing paired with high-resolution mass-spectrometry imaging (NanoSIMS) approaches. We have achieved our goals, publishing one high-impact paper on results from Objective 1; we expect to publish two more papers on results from Objective 2, described below.
Scientific Approach and Accomplishments
Objective 1: Characterize epiphyte diazotroph colonization and symbiosis maintenance using global gene-expression experiments
Current understanding of colonization and symbiosis maintenance in cyanobacteria-plant symbiosis at the molecular level is mostly based on studies of endophytes (reviewed in Santi et al. 2013). We sought to characterize gene acquisitions and regulatory rewiring that allowed epiphytic Nostoc to establish a symbiotic relationship with feather mosses. We hypothesized that Nostoc would (1) respond with a distinct gene expression profile to host mosses, (2) foster a distinct extracellular environment when in chemical contact with the moss, and (3) exhibit distinct metabolic processes and resource exchange compared to endophytes.
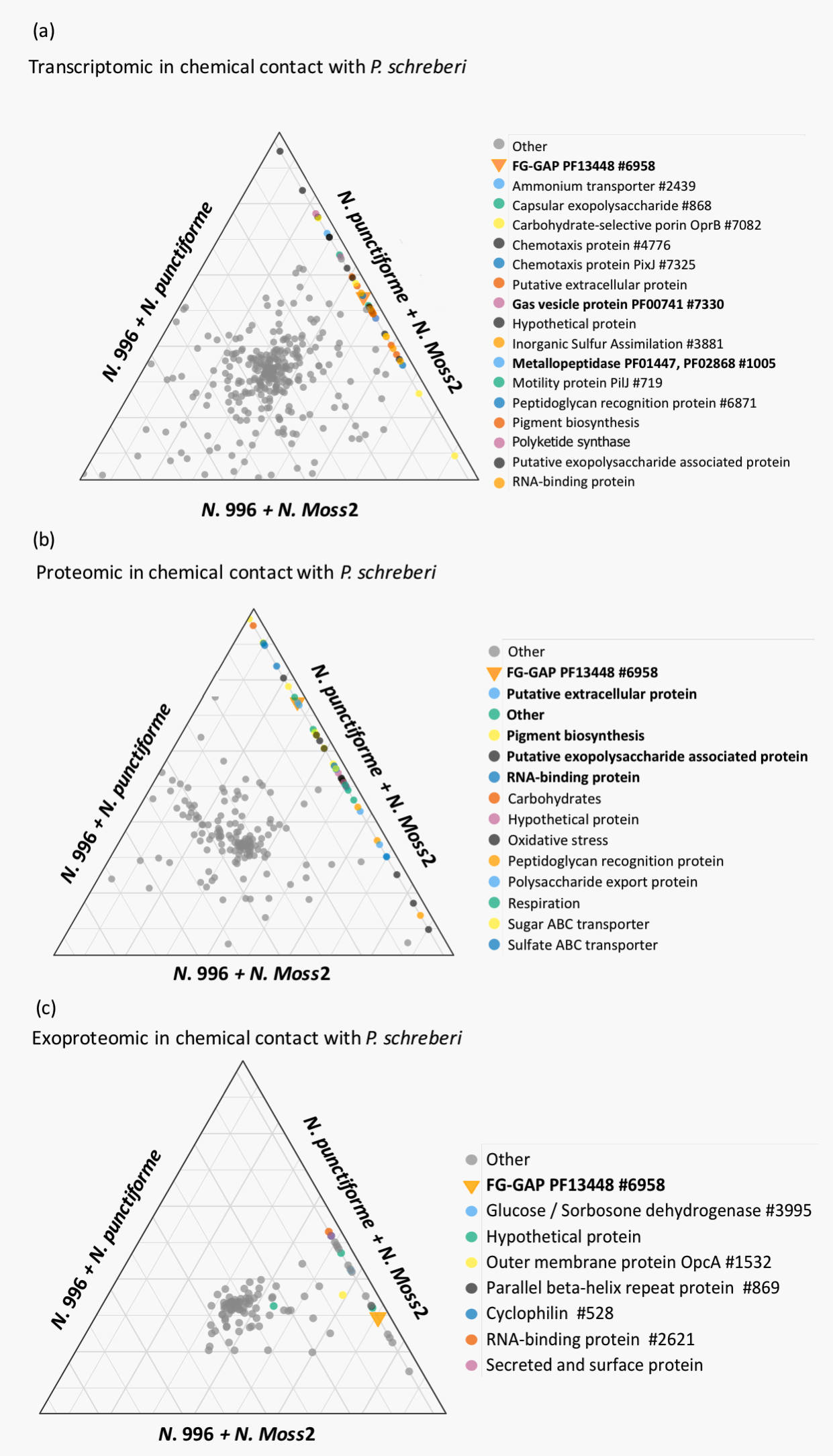
To test these hypotheses, we conducted a series of experiments investigating global gene, protein, and exoprotein expression of Nostoc strains that were able to form symbioses with the feather mosses ("symbiotic competent"), compared to a close relative that was unable to form a symbiotic relationship with the moss ("symbiotic incompetent”). Using permeable membranes, we designed an experimental setup that allowed us to distinguish between two phases of colonization: the chemical-signaling initiation phase and the physical-interaction phase.
Our resulting data provided evidence for the existence of a distinct suite of differentially expressed or newly acquired genes that the symbiotic-competent strains expressed while in contact with the moss, and a distinct extracellular environment (Figure 1). We were able to identify the genes required for this epiphytic symbiosis to occur through the competent-unique expression signatures at each of the two stages of colonization. These include several novel symbiosis-associated genes, such as sulfate transporters, and others identified in unrelated symbioses, such as nitric-oxide signaling. Exoprotein analyses uncovered symbiotic-competent-unique proteins, including a pectate lysase that may assist colonization.
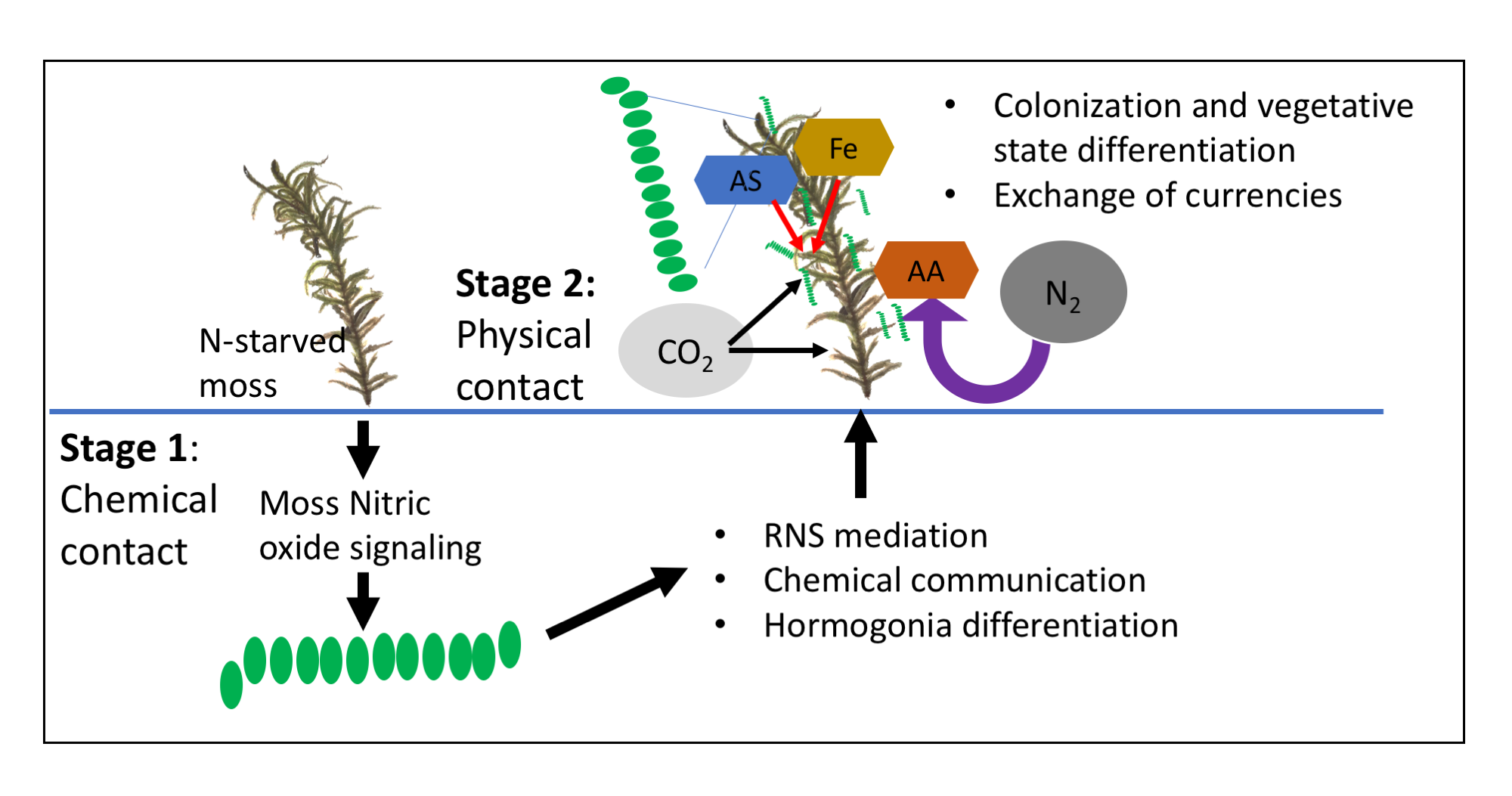
We also found evidence that this symbiosis is distinct from endophytic diazotroph symbioses. By examining changes in gene expression during the different stages of colonization, we were able to build a metabolic interaction model (Figure 2). Surprisingly, based on these expression analyses, sulfur and iron (most likely present as aliphatic sulfonates and ferritin-bound Fe, respectively) appear to be the major resources provided by the moss host, not organic C (as is found in endosymbionts). Additionally, fixed N is provided to the host as amino acids, not ammonium. The results of this work are presented in a publication in the ISME Journal (Warshan et al. 2017).
Objective 2: Use stable isotope tracing to test interaction model resource exchange
In N-fixing mutualistic interactions, exchange of resources is presumed but not often measured because specialized techniques such as NanoSIMS paired with stable-isotope probing are required. Results from Objective 1 indicated that S and Fe are the major resources provided by the plant host to the symbiont, which is novel. Sulfur is increasingly recognized as an important element for mediating interactions and signaling (Durham et al. 2015), but has not been explored in N-fixing symbionts. Iron is a co-factor of the nitrogenase enzyme, which performs N-fixation, so investigating Fe levels was also of interest to our research.
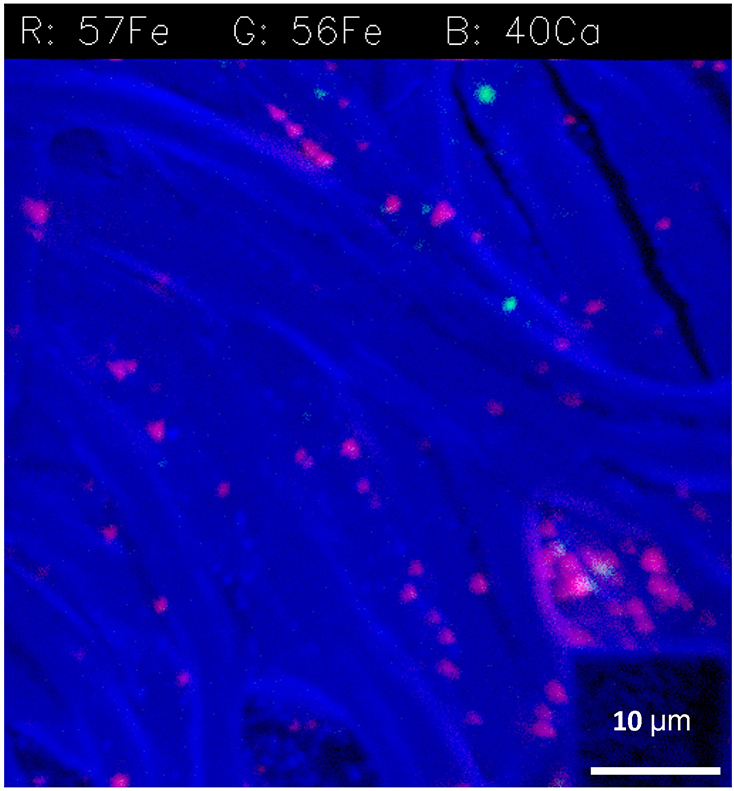
Iron transfer is particularly difficult to trace, since cellular concentrations can be very low. To address this, we developed methods for tracing Fe in diazotrophs using NanoSIMS and stable isotopes (Figure 3). There are four stable isotopes of Fe, including 56Fe, which is the most abundant in nature (91 percent), and the rarer isotopes 54Fe (5.8 percent), 57Fe (2.1 percent), and 58Fe (0.28 percent). We generated 57Fe-enriched minerals that we incubated with a model diazotroph that is often limited for Fe in its natural environment (Rubin et al. 2011). Using this approach, we were able to distinguish added stable isotopes from 56Fe particles present in the diazotroph colony prior to incubation (Figure 3).
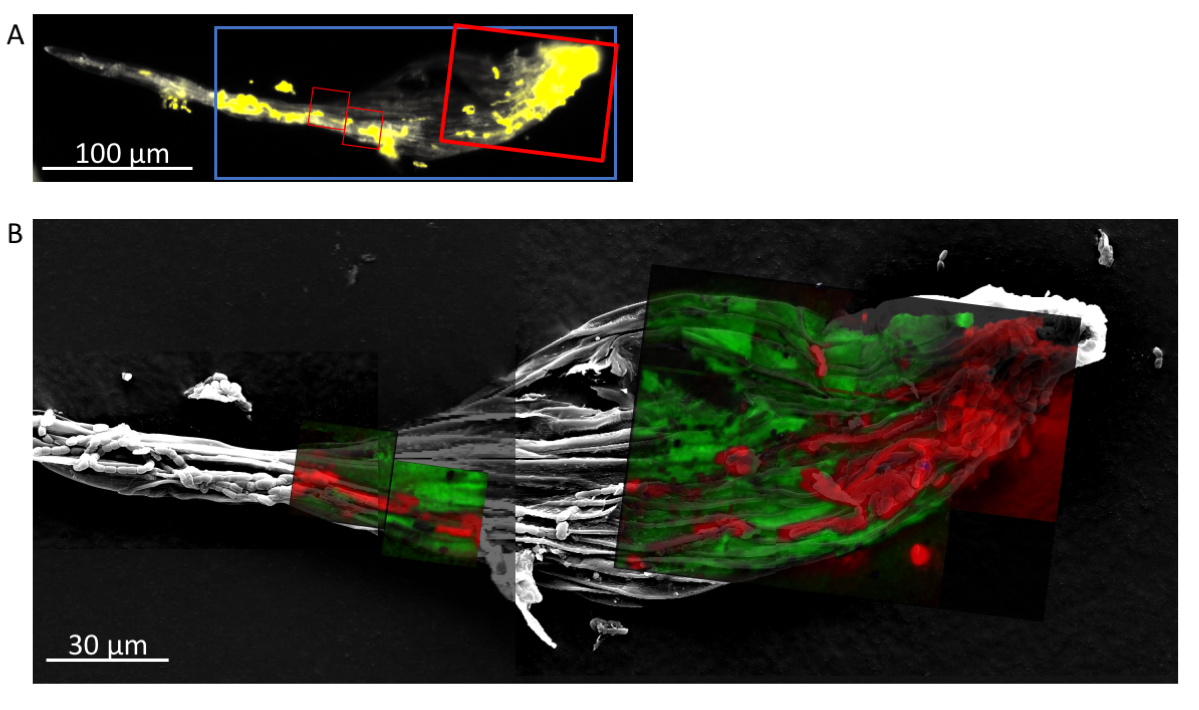
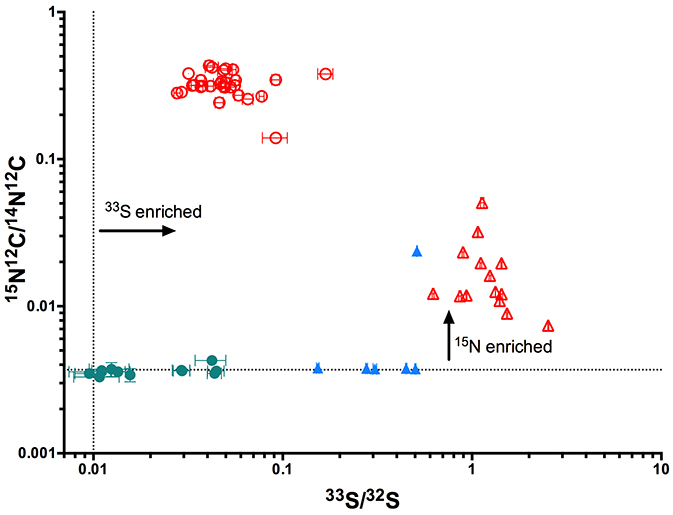
These new methods of tracing Fe in diazotrophs provided a path forward to test our interaction model (outlined in Figure 2). We used stable-isotope tracing to examine uptake and exchange of S, N, and C between partners. We generated moss hosts that were labeled with the rare stable isotopes for S, Fe, and C (33S, 57Fe, and 13C). This was accomplished by growing the moss with the rare stable isotope substituted in the medium for six months and several rounds of growth, the final round of which was in N-depleted medium in order to starve the moss for N. In order to trace transfer of the 33S, 57Fe, and 13C resources from the moss to the Nostoc, we set up colonization and incubation with unlabeled Nostoc symbionts. Additionally, to quantify N-fixation and transfer from the Nostoc to the moss, incubations were conducted in chambers with 15N2 gas (15N is the rare stable isotope of N). We then quantified 33S, 13C, and 15N isotope incorporation into the symbiont and moss tissue using NanoSIMS. The isotopes 33S and 13C in Nostoc cells would indicate transfer from the moss, and 15N in the moss would indicate N-transfer from Nostoc to the moss. Figure 4 depicts a representative NanoSIMS analysis of a moss leaf (phyllid) colonized by Nostoc. Nostoc is visible as a red color because of its high 15N levels due to N-fixation; moss tissue is green due to high initial 33S enrichment.
Following a 12-day incubation period, low but significant levels of 33S were detectable in the Nostoc that had colonized the moss (Figure 5). This is the first time the transfer of S has been demonstrated in cyanobacteria-plant symbioses. We also detected low levels of 15N in the moss tissue, validating the N-exchange that drives the symbiosis. This was demonstrated by other researchers (Bay et al. 2013), but it can now be linked to other element-exchange rates. The 13C isotope also appeared to have been transferred from the moss to Nostoc cells, but to confirm this, earlier time-points and additional controls would be required. Regardless, we have demonstrated S-transfer from the moss host to the cyanobacterial symbiont, which is novel.
Impact on Mission
This research supports Lawrence Livermore National Laboratory's core competencies of bioengineering and advanced energy systems, as well as its energy-security focus area. Our research has also contributed to the Laboratory's capabilities for studying the biological exchange of metals combining NanoSIMS imaging with molecular and biochemical characterization of metabolic pathways. Our work supports the DOE's goals of timely, material, and efficient transformation of the nation’s energy system and to secure U.S. leadership in clean-energy technologies. Our research has also contributed to DOE's and the Laboratory's carbon-capture-and-storage knowledge base: the new model system we developed and the related endophytic Sphagnum moss-Nostoc symbiosis represent the major nitrogen inputs to nitrogen-limited boreal ecosystems, which store up to 30 percent of terrestrial carbon (Lindo et al. 2013).
Conclusion
Biological N-fixation is carried out by some species of microbes and has the potential to eliminate the need for N fertilization; however, we are not able to apply these tools because we cannot adequately apply the relevant organisms to achieve N-fertilizer-free crop management. We established a novel model system of a promising epiphytic diazotroph symbiosis and used it to define genes, metabolic pathways, and putative resources exchanged between partners. Our results show that this diazotroph is surprisingly independent from its host. These findings provide a unique opportunity to perturb this system in a targeted manner and gain a mechanistic understanding of the interaction. One possible application of this research is to begin manipulation of the symbioses to transfer to novel plant hosts to enable bioenergy crop production.
While this research focuses on N-fixation applications for sustainable biofuel production, a basic mechanistic understanding of microbial interactions and response to a host is also broadly applicable. For example, biomedical research lacks the techniques to understand and predict mutualistic behavior between beneficial microorganisms in our bodies in response to disease (Faust and Raes 2012). This research has demonstrated a broadly applicable approach that we are uniquely suited to continue applying at Lawrence Livermore National Laboratory, due to our ability to combine NanoSIMS imaging of biological exchanges of carbon, nitrogen and iron, with molecular and biochemical characterization of metabolic pathways.
References
Andrews, M., et al. 2003. "Use of Nitrogen Fixing Bacteria Inoculants as a Substitute for Nitrogen Fertilizer for Dryland Graminaceous Crops: Progress Made, Mechanisms of Action and Future Potential." Symbiosis 35 (1):209—229.
Bay, G., et al. 2013. "Boreal feather mosses secrete chemical signals to gain nitrogen." New Phytologist 200 (1):54—60. doi: 10.1111/nph.12403.
Berrabah, F., et al. 2015. "Multiple Steps Control Immunity During the Intracellular Accommodation of Rhizobia." Journal of Experimental Botany 66 (7):1977-1985. doi: 10.1093/jxb/eru545.
Cocking, E. C., et al. 2012. "From Plant Protoplasts to Diazoplasts: The Challenge of Establishing Symbiotic Nitrogen Fixation in Cereals." Science and Culture 78 (5–6).
Congress, U.S. 110th. 2007. "Energy Independence and Security Act of 2007." (Public Law):110–140; http://www.gpo.gov/fdsys/pkg/PLAW-110publ140/pdf/PLAW-110publ140.pdf.
DeLuca, T. H., et al. 2008. "Ecosystem Feedbacks and Nitrogen Fixation in Boreal Forests." Science 320 (5880):1181–1181. doi: 10.1126/science.1154836.
——— 2002. "Quantifying Nitrogen-Fixation in Feather Moss Carpets of Boreal Forests." Nature 419 (6910):917–920. doi: 10.1038/nature01051.
Diaz, R. J., and R. Rosenberg. 2008. "Spreading Dead Zones and Consequences for Marine Ecosystems." Science 321 (5891):926–929. doi: 10.1126/science.1156401.
DOE, U.S. 2014. "Research for Sustainable Bioenergy: Linking Genomic and Ecosystem Sciences, Report from the October 2013 Workshop, DOE/SC-0167." U.S. Department of Energy Office of Science.
Durham, B. P., et al. 2015. "Cryptic Carbon and Sulfur Cycling Between Surface Ocean Plankton." Proceedings of the National Academy of Sciences of the United States of America 112 (2):453–457. doi: 10.1073/pnas.1413137112.
Faust, K., and J. Raes. 2012. "Microbial Interactions: From Networks to Models." National Review of Microbiology 10 (8):538-50. doi: 10.1038/nrmicro2832.
Gelfand, I., et al. 2013. "Sustainable Bioenergy Production from Marginal Lands in the U.S. Midwest." Nature 493 (7433):514–517. doi: 10.1038/nature11811.
Lindo, Z., et al. 2013. "Bryophyte-Cyanobacteria Associations as Regulators of the Northern Latitude Carbon Balance in Response to Global Change." Global Change Biology 19 (7):2022–2035. doi: 10.1111/gcb.12175.
Rubin, Maxim, et al. 2011. "Dust- and Mineral-Iron Utilization by the Marine Dinitrogen-Fixer Trichodesmium." Nature Geoscience 4 (8):529–534. doi: 10.1038/NGEO1181.
Santi, C., et al. 2013. "Biological Nitrogen Fixation in Non-Legume Plants." Annals of Botany 111 (5):743–767. doi: 10.1093/aob/mct048.
Stokstad, E. 2016. "The Nitrogen Fix." Science 353 (6305):1225–1227. doi: 10.1126/science.353.6305.1225.
Tilman, D., et al. 2006. "Carbon-Negative Biofuels from Low-Input High-Diversity Grassland Biomass." Science 314 (5805):1598–1600. doi: 10.1126/science.1133306.
Valentine, J., et al. 2012. "Food vs. Fuel: The Use of Land for Lignocellulosic ‘Next Generation’ Energy Crops that Minimize Competition with Primary Food Production." Global Change Biology Bioenergy 4 (1):1–19. doi: 10.1111/j.1757-1707.2011.01111.x.
Warshan, Denis, et al. 2017. "Feathermoss and Epiphytic Nostoc Cooperate Differently: Expanding the Spectrum of Plant-Cyanobacteria Symbiosis." ISME Journal 1–13. doi: 10.1038/ismej.2017.134.
Weston, D. J., et al. 2015. "Sphagnum Physiology in the Context of Changing Climate: Emergent Influences of Genomics, Modeling and Host-Microbiome Interactions on Understanding Ecosystem Function." Plant, Cell & Environment 38 (9):1737–1751. doi: 10.1111/pce.12458.
Publications and Presentations
Warshan, Denis, et al. 2017. "Feathermoss and epiphytic Nostoc cooperate differently: expanding the spectrum of plant-cyanobacteria symbiosis." ISME J. doi: 10.1038/ismej.2017.134. LLNL-JRNL-730832.
Stuart, R. K. 2016. “Tracing Microbial Community Utilization of Light-Derived Extracellular Organics.” Lawrence Livermore National Laboratory Biosciences and Biotechnology Division Seminar, Livermore, CA. LLNL-PRES-708697.
Stuart, R. K., et al. 2016a. “Characterizing Extracellular Functions in Diverse N-Fixing Cyanobacteria.” American Geophysical Union Annual Meeting, San Francisco, CA. LLNL-ABS-700218.
——— 2016b. “Characterizing extracellular functions in diverse N-fixing cyanobacteria.” American Geophysical Union Annual meeting, San Francisco, CA. LLNL-POST-713518.