Peter Barnes (14-ERD-062)
Abstract
The nation faces more and more problems that are global in scale, especially in the field of national security, yet lacks the capability of simulating those problems at worldwide scale to help develop the best possible countermeasures. We intend to develop global-scale models to predict outcomes, evaluate courses of action, and develop new analysis techniques for addressing global problems. To demonstrate the capabilities of our global modeling approach, we will develop applications based on discrete-event simulation, which models the operation of a system as a discrete sequence of events in time, with each event occurring at a particular instant and marking a change of state in the system. For this project, we will model a distributed denial-of-service cyber attack as well as a pandemic model that simulates the spread of an evolving virus. We will tackle three fundamental research challenges in discrete-event simulation: (1) the automatic reverse engineering of source code; (2) dynamic load balancing, a technique that distributes processing equally among processors throughout the job's life; and (3) global synchronization using only one-sided communication data.
Using Sequoia-class supercomputers, we expect to develop and implement planetary-scale models such as a real-time model with an agent for every person on earth, executing five events per second per person; a model of the entire Internet, transmitting 10 packets per second per node; or a worldwide pandemic model simulating viral evolution during replication and transmission. We will aim for planetary scale, and in so doing learn a tremendous amount about the underlying technical challenges.
Mission Relevance
This project supports the Laboratory's strategic focus area in cyber security, space, and intelligence, and core competency in high-performance computing, simulation, and data science by developing a new, scalable discrete-event simulation system that will support the predictive modeling of complex systems critical to national security. This project also supports LLNL's bioscience and bioengineering core competency by advancing computational biology tools for outcome prediction, especially using genomic sequences, related protein sequences, and protein structure models to help predict how viruses might evolve.
FY15 Accomplishments and Results
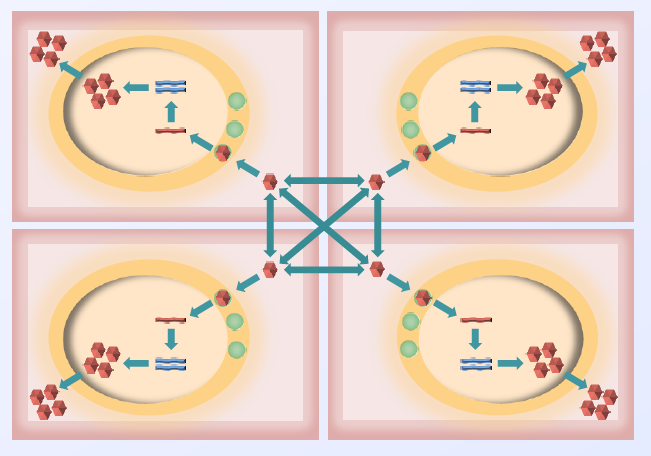
In FY15 we (1) generated the Backstroke optimistic code, an open-source reverse-code generation framework; (2) released a beta version for testing by the Georgia Institute of Technology, Atlanta; (3) completed the coupling of ROSS+ (Rensselaers' optimistic simulation system) and the object-oriented parallel run-time system Charm++; (4) developed a fully asynchronous global virtual-time algorithm; (5) implemented dynamic load balancing; (6) refined the implementation of the viral evolution model (see figure); (7) evaluated the feasibility of supporting the ns-3 simulator, an open-source discrete-event network simulator for Internet systems, in an optimistic parallel discrete-event simulation engine; and (8) developed a demonstration model of securities markets to show the utility and scaling of parallel discrete-event simulation engines for policy analysis.
Publications and Presentations
- Nikolaev, S., et al., “Pushing the envelope in distributed ns-3 simulations: One billion nodes.” WNS3 '15: Proc. 2015 Workshop on ns-3, ACM, New York, NY, p. 67 (2015). LLNL-CONF-667630.
- Smith, S. G., et al., Improving per processor memory use of ns-3 to enable large scale simulations. Workshop on ns-3 (WNS3), Barcelona, Spain, May 13–14, 2015. LLNL-CONF-667822.