Rhona Stuart (16-LW-030)
Project Description
A national mandate requires that about 25% of the country's liquid transportation fuel must come from "nongrain biofuels" by 2022. The feedstock for these types of fuels includes grasses, algae, or dry plant matter such as straw and forestry residues. This mandate could be achieved by high-productivity crops on fertile land, but at the expense of the nation's food supply. Growing biofuels crops on marginal lands is the best option for biofuel production that does not interfere with food production, but nitrogen fertilization, which would be required to help grow such feedstocks, is a major barrier to sustainable practices. Potential problems with using nitrogen fertilization include an increased nitrous oxide flux, which can negate the greenhouse-gas benefits of these fuels, create widespread damage to coastal ecosystems and their fisheries through nitrogen runoff, and add expense that cuts into the low profit margin of biofuel production. One possible solution to this problem is biological nitrogen fixation, a process in which nitrogen gas from the atmosphere is incorporated into the tissue of certain plants with the help of soil microorganisms that live in close association with their plant hosts. However, translating the natural capabilities of these nitrogen-fixing microorganisms to high-productivity monoculture crop environments is a challenge. For example, microorganisms that are known to fix nitrogen in the environment do not always fix nitrogen in a culture, or if they do, the rates are lower than in the field. Helper (i.e., probiotic) bacteria can increase nitrogen-fixation rates, but the mechanisms by which they do so are unknown. We plan to use molecular biology and isotope tracing to identify the symbiotic mechanism, called mutualism, between nitrogen-fixing microorganisms and a helper bacterium. Once we understand the mechanism, we can apply the knowledge to promote sustainable biofuel crop growth to meet the national mandate. In addition, the work from this project will also demonstrate a new combination of tools that will be widely applicable to studies of interactions in microbial communities, such as the those within the human body.
Results from this project will provide us, for the first time, with a mechanistic understanding of a nitrogen-fixing bacterial mutualism, quantifying exchange and expression changes under relevant conditions. Results will include the identification of biomarkers and conditions necessary for future manipulation of a valuable microbial mutualism to enable sustainable biofuel production. A basic understanding of the triggers and drivers of this association will enable engineering and manipulation of similar pairs of microbes that could be applied to production of biofuel crops such as switchgrass. On its own, mapping the exchange of carbon and nitrogen under variable conditions between mutualistic bacteria will substantially advance the field of microbial community studies. Mapping iron exchange between bacteria using stable isotopes, which has never been done, has the potential to open up a new area of nutrient–metal exchange studies in this field. Combining stable isotope approaches with physiological experiments and gene-expression profiling addresses a key gap in microbial studies by defining mechanisms of how and why microbes benefit one another. Thus, our work will provide a tool that can be used to predict microbial interactions in complex microbial communities as diverse as the human body and the climate, a key goal for future microbial engineering in biofuel and biomedical research.
Mission Relevance
This project combines LLNL strengths in high-resolution imaging and complex microbial community analyses, and is directly related to ongoing Livermore research in biotechnology, microbial nutrient cycling, carbon sequestration, and climate change. Such work supports the Laboratory's core competency in bioscience and bioengineering. The development of new tools to investigate the uptake and exchange of carbon, nitrogen, and iron will also enhance Livermore's nuclear, chemical, and isotopic science and technology core competency by promoting the continued development and early application of high-resolution imaging mass spectrometry.
FY16 Accomplishments and Results
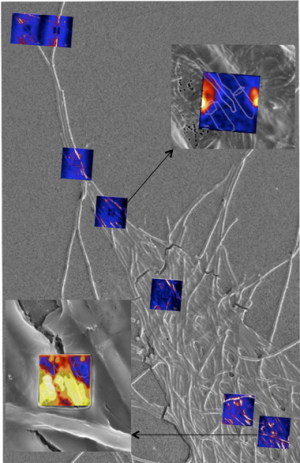
In FY16 we (1) identified the secreted proteome complement of proteins in nitrogen-fixing cyanobacteria in symbiosis with moss and helper bacteria—these proteins were identified under variable conditions of contact to identify mutualistic behavior between the moss, helper bacteria, and several related strains of nitrogen-fixing cyanobacteria; (2) validated pathways of mutualism that may involve both sulfur and iron exchange between the microbes and the moss; and (3) conducted experiments to examine iron acquisition and exchange in a nitrogen-fixing cyanobacterium using iron isotopes and nanoscale secondary-ion mass spectrometry, with iron from variable sources (hematite, ferrihydrite, and bacteria), which are used to trace iron into both the nitrogen-fixing cyanobacteria and surrounding bacteria (see figure).