Anne Dekas (13-LW-032)
Abstract
Thaumarchaea, a recently recognized archaeal phylum, comprise over 20% of the total microbial cells in marine waters. However, their activity and metabolic flexibility are poorly described. We sought to investigate the ability of these archaea organisms to assimilate a variety of carbon sources, expanding their known role in the marine carbon cycle. We incubated coastal Pacific Ocean water in the dark with substrates labeled with carbon-13, nitrogen-15, or both carbon-13 and nitrogen-15. Using nanoscale secondary-ion mass spectrometry and automated particle-recognition software, we analyzed over 5,000 individual cells. After 7 days, 95 and 89% of total microbial cells (from deep and shallow depths, respectively) demonstrated anabolic activity—that is, incorporation of at least one isotopically labeled substrate. Chemoautotrophy was detected at both sites, with 36 and 9% of cells (deep and shallow, respectively) assimilating bicarbonate labeled with carbon-13 in the dark. Fluorescence in situ hybridization coupled to nanoscale secondary-ion mass spectrometry analysis was performed to link 16S rRNA phylogeny to patterns of carbon assimilation. Thaumarchaea dominated chemoautotrophy, with carbon-13 bicarbonate assimilation in nearly all Thaumarchaeal cells analyzed and none detected in bacterial cells. Thaumarchaeal cells were enriched in nitrogen-15 after incubation with carbon-13 and nitrogen-15 amino acids, but not carbon-13, suggesting selective nitrogen assimilation from amino acids or substrate recycling. Together, our results demonstrate the value of single-cell measurements in characterizing patterns of carbon metabolism in mixed microbial communities, and underscore the importance of Thaumarchaea in marine chemoautotrophy. Future analyses of the samples collected will continue to shed light on the carbon assimilatory capabilities of Thaumarchaea.
Background and Research Objectives
Microbial life is central to the global carbon and nitrogen cycles, but its major players and metabolic diversity are still being identified and characterized. Using experiments and technological development, we used soil and marine samples to determine the carbon and nitrogen metabolism of Thaumarchaeota (“thaumas” from the Greek “wonder”), a ubiquitous microbial cell in marine waters that likely plays an important role in marine biogeochemical cycles.1 Thaumarchaeota are thought to live primarily chemoautotrophically, coupling ammonia oxidation to bicarbonate fixation. Some evidence suggests that they may live heterotrophically, obtaining food and energy by taking in organic substances, or mixotrophically, that is existing either heterotrophically or autotrophically (using inorganic materials and photosynthesis or chemosynthesis).2–5 This possibility has implications for the rates at which these cells process greenhouse gases, particularly carbon dioxide and nitrous oxide. Thaumarchaeota are thought to constitute a significant carbon dioxide sink, but also have been shown to contribute significantly to atmospheric nitrous oxide, a greenhouse gas. Previous attempts to investigate their metabolic capabilities have largely depended on their isolation and individual physiological characterization, a slow process that yields an incomplete view of the activity of these microorganisms in situ. To date, fewer than 15 strains of the Thaumarchaeota (marine or terrestrial) have been obtained in pure culture or enrichment.6 Culture-independent techniques, such as gene sequencing, have therefore been critical to developing our understanding of the distribution and phylogenetic diversity of these organisms.7,8 Still, we have a poor understanding of their metabolic capabilities and activities in situ, and therefore their contribution to biogeochemical cycling.
The potential carbon assimilatory strategies of marine Thaumarchaea have different implications for their production and consumption of greenhouse gases. All currently cultured representatives of Thaumarchaea are chemoautotrophic, coupling bicarbonate fixation with ammonia oxidation. However, the metabolic capabilities of these diverse organisms in situ is likely more complex. Evidence of Thaumarchaeal autotrophy includes the presence of the genes necessary for a modified 3-hydroxypropionate/4-hydroxybutyrate cycle,4 and the ability of some members to assimilate bicarbonate.9,10 However, genes encoding amino acid transporters and the oxidative tricarboxylic acid cycle as well as the ability of some members to assimilate organic matter labelled with hydrogen-3 or carbon-13 provide evidence for Thaumarchaeal heterotrophy.4,11,12 Several researchers have proposed that Thaumarchaeota live mixotrophically, supplementing a primarily bicarbonate diet with organic carbon sources.6,10,13,14 Still other research suggests pure heterotrophy or a lack of ammonia oxidation.11,15 If the majority of Thaumarchaea live autotrophically while oxidizing ammonia, they act as a sink of carbon dioxide, but also as a source of nitrous oxide (a potent greenhouse gas and by-product of ammonia oxidation). However, if they live heterotrophically, they are a source of carbon dioxide, actively reducing the amount of organic carbon reaching the seafloor for ultimate burial.
The goal of our work was to investigate the carbon assimilatory capabilities of marine Thaumarchaea to better understand the role of these cells in global carbon and nitrogen cycling. To this end, we sought to (1) collect environmental samples containing Thaumarchaea, (2) characterize the microbial community and enrich for archaea using bacterial antibiotics, (3) determine if relative archaeal abundance or activity (transcripts and rate measurements) changed with the addition of various carbon sources, and (4) directly observe and characterize Thaumarchaeal carbon assimilation on the single-cell level.
Scientific Approach and Accomplishments
Sampling Sites
As shown in Figure 1, samples were collected at two sites in the Pacific Ocean. The first was 17 km off the coast of Los Angeles, California, in the San Pedro Basin (+33° 33’ 00, -118° 24’ 00) at the site of the San Pedro Ocean Time-series (SPOT), which is is a long-term study of core biogeochemical measurements. The second site was 0.4 km off the coast of western San Francisco, California, at the end of Pacifica Pier. A circular rosette of sampling Niskin bottles was used to collect the San Pedro Basin samples at 150-m water depth. A line and bucket was used to collect the Pacifica Pier samples from the surface of the water. In total, over 1,000 L of water were collected. On the two San Pedro Basin trips, approximately 10 L of water were filtered onboard onto 0.2-µm polycarbonate filters and stored on dry ice for 16S rRNA gene and transcript analysis. Approximately 150 additional liters of water were filtered onboard the second sampling trip for meta-genomic and meta-transcriptomic analysis. The rest was transported to the laboratory for use in stable-isotope probing experiments.
Manipulation Experiments
Five sets of bottle incubations were performed. Seawater from the Pacifica Pier and the San Pedro Basin was incubated with a variety of substrates labeled with carbon-13 (bicarbonate, glucose, oxaloacetate, pyruvate, or lipids), substrates labeled with nitrogen-15 (ammonium ion, protein, urea, or amino acids) and/or doubly labeled substrates (carbon-13 and carbon-15 amino acids, urea, or protein). The water was incubated in acid-rinsed polycarbonate bottles (2, 4, or 11 L) in the dark at 4 to 10ºC for up to 7 days. At the completion of the experiment, the water was filtered with 0.2-µm polycarbonate filters and fixed with 2% paraformaldehyde (for microscope analysis), 0.2-µm polyethersulfone filters and frozen (for 16S rRNA gene and transcript analysis), and pre-combusted grade GF/F filters (for isotope-ratio mass spectrometry analysis).- Experiments 1 to 3 were performed to test the ability of individual Thaumarchaeal cells to assimilate inorganic carbon (carbon-13 bicarbonate) and/or nitrogen-15 amino acids, protein, and urea, and hence indicate mixotrophy. These experiments included treatments amended with bacterial antibiotics to increase the relative abundance of the archaeal population.
- Experiment 4 was performed to test the ability of Thaumarchaea to assimilate a wide variety of carbon substrates labeled with carbon-13, including glucose, lipids, amino acids, oxaloacetate, pyruvate, protein, urea, and bicarbonate.
- Experiment 5 was designed to measure community-level rates of organic carbon and ammonium uptake, as well as nitrification rates, and to assess whether the addition of organic carbon affected nitrification rates.
Microbial Community Characterization
The composition of the microbial communities was characterized by sequencing the 16S rRNA genes and transcripts extracted from both the t0 and tfinal samples. The DNA and RNA were extracted from frozen filters, RNA was converted to cDNA, and the 16S rRNA gene was amplified using primers targeting the V4 and V5 regions, 515F and 926R, with a modification.16 Illumina tag sequencing was performed at the University of California, Davis, using a MiSeq sequencer system. Sequences were trimmed using PRINSEQ, which is a software used to filter, reformat, or trim genomic and meta-genomic sequence data. We then merged the data with USEARCH7 (used for high-throughput biological sequence analysis), using a 99% cut-off for operational taxonomic unit clustering (an operational definition of a species or group of species often used when only DNA sequence data is available). Finally, the data was de-multiplexed in QIIME, an open-source bioinformatics pipeline for performing microbiome analysis from raw DNA sequencing data. The DNA and RNA for meta-genomic and meta-transcriptic analysis was also extracted and sequenced at the University of California, Davis, using an Illumina HiSeq sequencing platform.
Community Composition and Bottle Effects
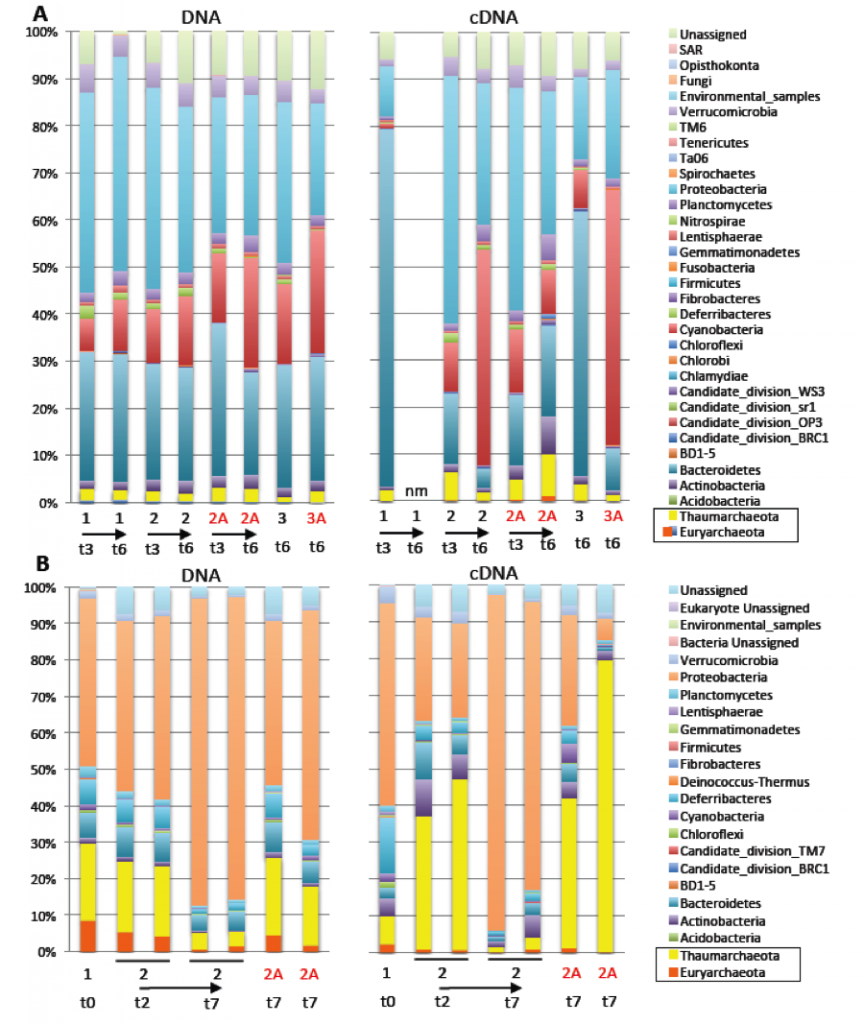
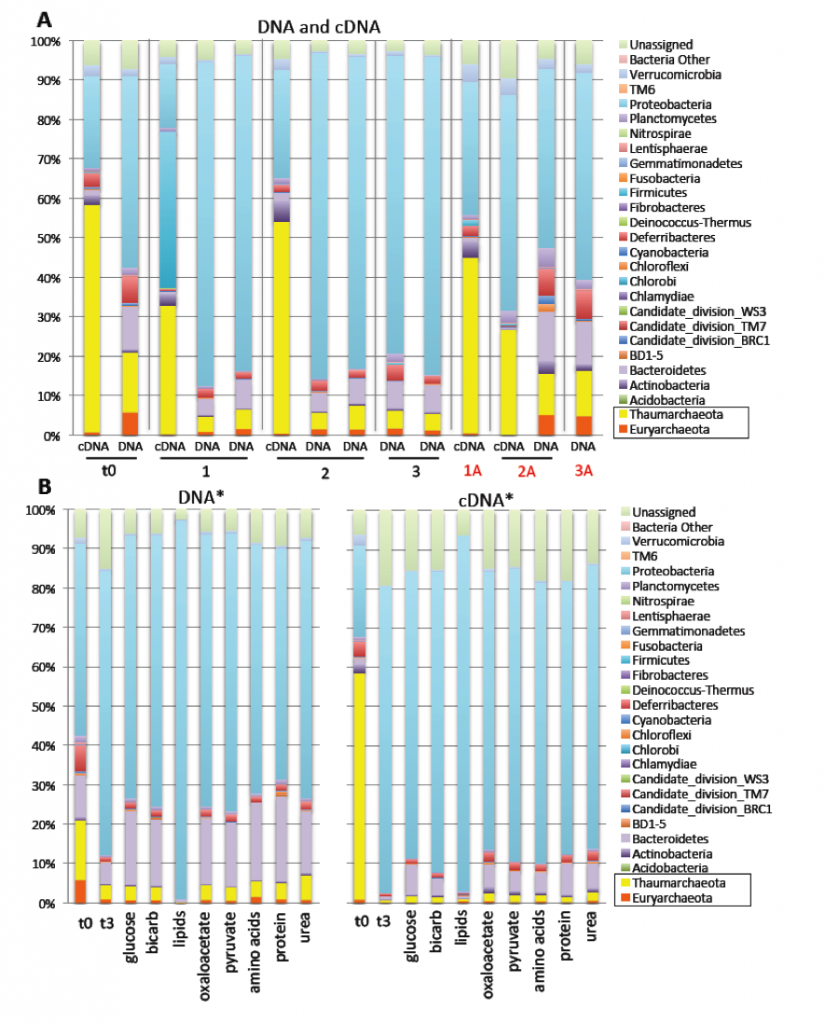
We successfully characterized the composition (DNA) and activity (cDNA) of the microbial communities at our two sampling sites, as well as over the course of our manipulation experiments (Figures 2 and 3). Thaumarchaea and Euryarchaea were relatively abundant at the San Pedro Basin, comprising 21 and 8% of the total in situ microbial community during the first sampling trip, and 15 and 6% during the second. Proteobacteria were the most abundant bacterial phylum. In both experiment 2 and experiment 5, the archaeal community decreased in relative abundance and activity over time in the incubation bottles. Somewhat surprisingly, because archaea are thought to be relatively slow growing in marine waters, Thaumarchaeal transcripts dominated the community during the second San Pedro Basin trip. The initial community composition was not characterized at the Pacifica Pier, but after 3 days of incubation with no substrates added, the Thaumarchaea comprised 2.4% of the community. Although they are typically relatively less abundant in shallow waters, their absolute abundance is constant with depth, because surface waters contain approximately an order of magnitude more cells per unit volume than water at 150 m (~106 cells/mL versus ~105 cells/mL).
Effect of Antibiotics on Community Composition
Addition of bacterial antibiotics (ampicillin and streptomycin) increased the relative abundance and activity of archaea in Pacifica Pier waters only minimally, but had a more substantial effect in San Pedro Basin waters. The biggest effect was observed during experiment 2, when antibiotics resulted in a Thaumarchaeal community of 18%, on average, versus 4.5% in the equivalent treatment without antibiotics. Additionally, Thaumarchaeal transcripts comprised 41 and 79% of the total with antibiotics, and only 1.5 and 3% without. Although the bacterial community was not completely inhibited, this suggests that bacterial antibiotics can effectively enrich for archaea even on short timescales.
Effect of Organic Carbon Additions on Community Composition
The isotopically labeled substrates were added at low (nanomolar) concentrations to prevent overgrowth of opportunistic heterotrophic bacteria, and therefore unnatural disruptions to the community structure. Experiment 5 demonstrated that most of the organic carbon substrates added (glucose, oxaloacetate, pyruvate, amino acids, and urea) were added at sufficiently low concentrations to not change the community composition or activity at the phylum level. Fertilizing effects were therefore successfully minimized in this experiment.
Community-Level Metabolic-Rate Measurements
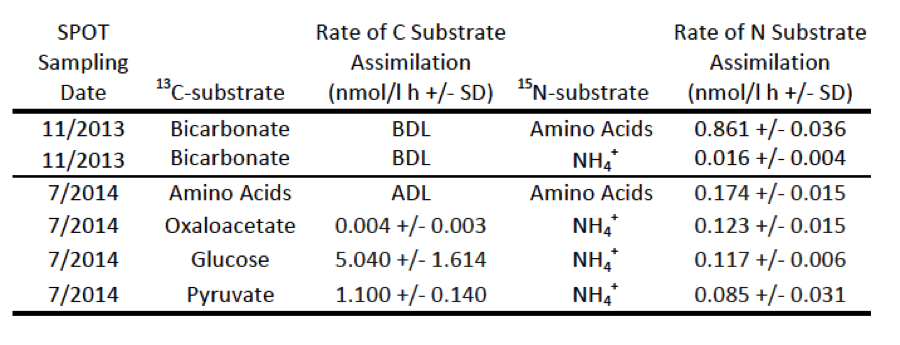
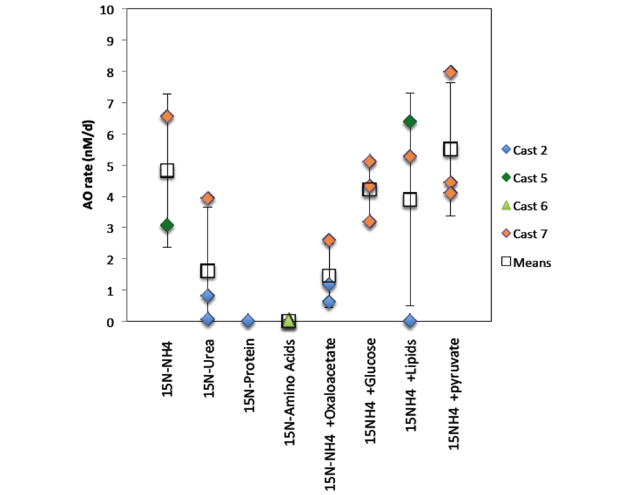
To understand the overall activity levels of the community at 150-m water depth, we measured bulk metabolic rates of both carbon-assimilation and nitrification (Table 1 and Figure 4). Carbon- and nitrogen-assimilation rates were calculated based on the incorporation of carbon-13 and nitrogen-15 substrates as measured by increasing carbon and nitrogen isotope ratios, measured by isotope-ratio mass spectrometry at the University of Southern California, Los Angeles, California. Nitrification rates were measured by isotopic analysis of nitrate, measured on an isotope-ratio mass spectrometer using the "denitrifier" method.17 Nitrification was observed, not only canonically with the NH4+ammonium ion (presumably converted to ammonia before oxidation) as the substrate, but also with urea. Rates of nitrification with the ammonium ion varied between incubations, but this is likely because of variations in microbial communities (water was collected in seven different efforts, or “casts,” over the course of the day) rather than the effect of the organic carbon substrates added.
Table 1. Community-level metabolic rates in San Pedro Basin samples (experiments 2 and 5) measured by isotope-ratio mass spectrometry. BDL = below detection limit, ADL = above detection limit, SD = standard deviation of biological triplicates.
Single-Cell Anabolic Activity Assessed by Nanoscale Secondary-Ion Mass Spectrometry
To determine the substrate assimilation of individual cells, we measured their isotopic composition with Livermore's CAMECA NanoSIMS 50. A cesium-ion primary ion beam (2–4 pA) with a nominal spot size of 100 to 200 nm was used to rastor over the cells. Four masses were collected (12C2−, 12C13C−, 14N12C−, and 15N12C−) using electron multipliers. Bacillus spores with known isotopic composition (previously analyzed by isotope-ratio mass spectrometry) were used as standards. Images were processed using L’Image software, including an automated particle-finder feature. To link the 16S rRNA phylogeny to the isotope composition, fluorescence in situ hybridization and catalyzed reporter deposition were employed before nanoscale secondary-ion mass spectrometry analysis for a subset of samples. We carried out the hybridization and deposition using the Cren537 HRP-probe to target Thaumarchaea and Eub338 to generally target bacteria.18–20
Isotopic Analysis of Unidentified Community Members
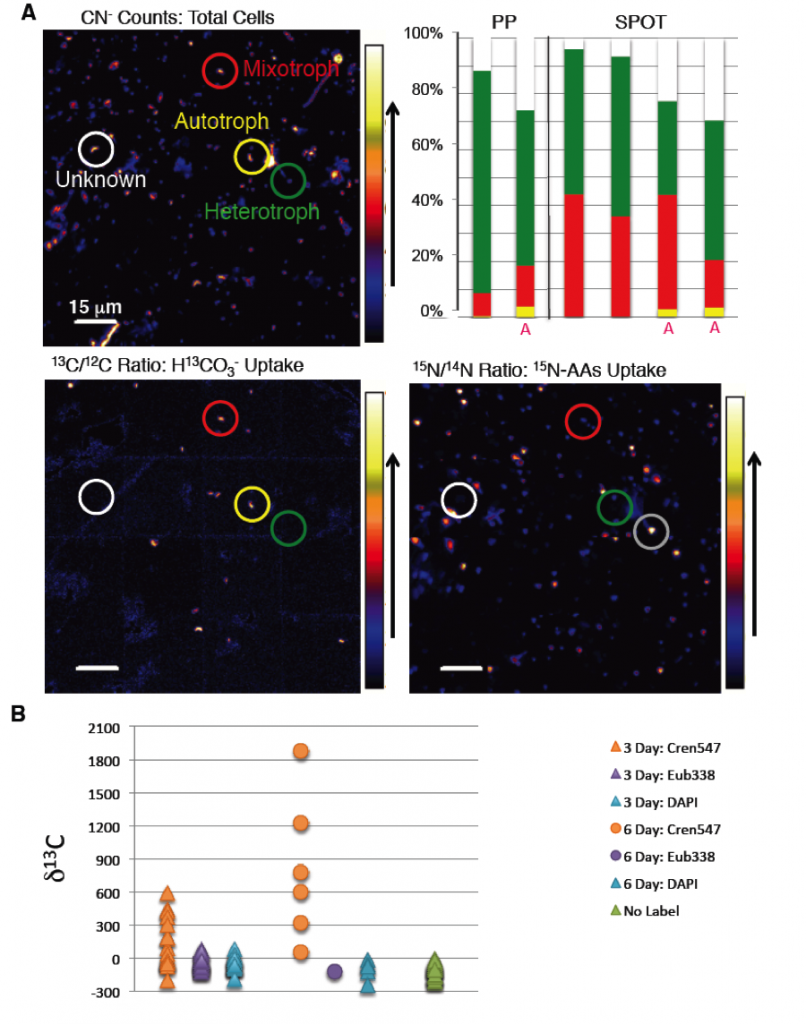
We analyzed the nitrogen-15 and nitrogen-14 as well as the carbon-13 and carbon-12 ratio of 4,968 individual cells incubated with carbon-13 bicarbonate, nitrogen-15 amino acids, or doubly labeled carbon-13 and nitrogen-15 amino acids, with and without antibiotics, from both the San Pedro Basin and Pacifica Pier (experiments 1 and 2). After 7 days, 95 and 89% of cells (San Pedro Basin and Pacifica Pier, respectively) demonstrated anabolic activity—that is, incorporation of at least one isotopically labeled substrate. Chemoautotrophy was detected at both sites, with 36 and 9% of cells (San Pedro Basin and Pacifica Pier, respectively) assimilating carbon-13 bicarbonate in the dark. The percentage of both autotrophs and mixotrophs was elevated, though only slightly, when treated with antibiotics in both sets of incubations. Combined with the sequencing results indicating that antibiotics enriched for archaea, this observation is consistent with archaea living both autotrophically and mixotrophically.
Isotopic Analysis of Phylogenetically Identified Cells
To specifically investigate the archaeal cells, we analyzed the isotopic composition of 50 Thaumarchaeal cells (identified by hybridization with the Cren537 probe), as well as 67 bacterial cells (identified by hybridization with the Eub338 probe), as shown in Figure 5. Thaumarchaea dominated chemoautotrophy at both San Pedro Basin and Pacifica Pier, with carbon-13 bicarbonate assimilation in nearly all Thaumarchaeal cells analyzed, but none of those hybridized with the general bacterial probe. Nitrogen-15 enrichment after incubation with nitrogen-15 amino acids was widespread in both the archaeal and bacterial populations. The co-localization of carbon-13 and nitrogen-15 uptake from inorganic and organic substrates suggested mixotrophy in the Thaumarchaea. However, the patterns of isotope incorporation during incubations of carbon-13 and nitrogen-15 amino acids challenged this interpretation. While bacteria were enriched in both carbon-13 and nitrogen-15 after incubation with the dual-labeled amino acids, the Thaumarchaeal cells were only enriched in nitrogen-15. Several possibilities may explain this, including preferential nitrogen incorporation from amino acids by Thaumarchaea or substrate recycling resulting in nitrogen-15 enrichment as a result of incorporation of the ammonium ion labeled with nitrogen-15. Additional analyses are required to differentiate these possibilities, but if the former is proven true, this is direct evidence for archaeal mixotrophy. The lack of detectable nitrification in the amino acid treatment of experiment 5 suggests that recycling of nitrogen-15 amino acids to nitrogen-15 ammonium ions was not a universally significant process, otherwise it would have been oxidized and detected as nitrification.
Geochemical Measurements
Nitrogen Measurements In Situ
Concentrations of ammonium and nitrite were measured at the University of Southern California, Los Angeles, California, in t0 water on both San Pedro Basin sampling trips. Both nitrogen species were below detection (<10 nM) in all samples measured on both trips, with the exception of nitrite in the last cast of water collected at 150-m depth in July 2014. This sample had 22-nM nitrite.
Delta Carbon-13 of Dissolved Inorganic Carbon
Delta carbon-13 is an isotopic signature, a measure of the ratio of stable isotopes carbon-13 to carbon-12, reported in parts per thousand. The delta carbon-13 of dissolved inorganic carbon of incubation filtrate was measured as a strontium carbonate ion precipitate by isotope-ratio mass spectrometry at the University of California, Davis. No elevation in the delta carbon-13 of dissolved inorganic carbon was observed over the course of the incubations (data not shown), consistent with calculations of the potential enrichment of the dissolved inorganic carbon pool assuming complete conversion of added dissolved organic carbon labeled with carbon-13 to carbon-13 dissolved inorganic carbon by heterotrophic metabolism. This validates the interpretation of carbon-13 enrichment during incubation with carbon-13 organic carbon substrates as the assimilation of organic matter and not an artifact of recycling.
Impact on Mission
This project is closely aligned with the Laboratory's strategic focus area of energy and climate security because of its potential to contribute to our understanding of global carbon cycling and modeling of the global carbon cycle. Determining carbon and nitrogen metabolism of new microorganisms directly aligns with the Laboratory’s core competency in bioscience and bioengineering.
Conclusion
We launched a multidisciplinary investigation of carbon cycling by a widespread and under-studied group of marine microorganisms, the Thaumarchaea. We successfully completed our goal of directly observing carbon anabolic activity in Thaumarchaea from two marine locations. We found that Thaumarchaea assimilate inorganic carbon in both shallow (surface) and deep (150-m) marine water. We additionally observed Thaumarchaeal uptake of nitrogen-15 but not carbon-13 when incubated with nitrogen-15 and carbon-13 amino acids, in contrast to the pattern of dual uptake observed in bacterial cells. This suggests either preferential assimilation of nitrogen from amino acids or substrate recycling during the experiment resulting in assimilation of the ammonium ion labeled with nitrogen-15. Our experiments with a range of organic carbon sources revealed that low levels of organic carbon additions do not affect the relative abundance or activity of marine archaea nor the rates of nitrification (presumably mediated by Thaumarchaea) over the short timescales investigated. Analyses of the samples collected during these experiments will continue to reveal clues about archaeal carbon cycling in the marine environment. Additional measurements with fluorescence in situ hybridization and nanoscale secondary-ion mass spectrometry and bioinformatics analyses have been planned. Our results demonstrate the value of single-cell measurements in characterizing patterns of carbon metabolism in mixed microbial communities, and underscore the importance of Thaumarchaea in marine chemoautotrophy.
References
- Karner, M. B., E. F. DeLong, and D. M. Karl, "Archaeal dominance in the mesopelagic zone of the Pacific Ocean." Nature 409, 507 (2001).
- Ouverney, C. C., and J. A. Fuhrman, "Marine planktonic archaea take up amino acids." Appl. Environ. Microbiol. 66(11), 4829 (2000).
- Konneke, M., et al., "Isolation of an autotrophic ammonia-oxidizing marine archaeon." Nature 437, 543 (2005).
- Hallam, S. J., et al., "Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota." PLoS Biol. 4(12), e437 (2006).
- Ingalls, A. E., et al., "Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon." Proc. Natl. Acad. Sci. Unit. States Am. 103(17), 6442 (2006).
- Hatzenpichler, R., "Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea." Appl. Environ. Microbiol. 78(21), 7501 (2012).
- Church, M. J., et. al., "Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean." Environ. Microbiol. 12(3), 679 (2010).
- Francis, C. A., et al., "Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean." Proc. Natl. Acad. Sci. Unit. States Am. 102(41), 14683 (2005).
- Pratscher, J., M. G. Dumont, and R. Conrad, "Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil." Proc. Natl. Acad. Sci. Unit. States Am. 108(10), 4170 (2011).
- Ingalls, A. E., et al., "Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon." Proc. Natl. Acad. Sci. Unit. States Am. 103(17), 6442 (2006).
- Seyler, L. M., L. M. McGuinness, and L. J. Kerkhof, "Crenarchaeal heterotrophy in salt marsh sediments." ISME J. 8(7), 1534 (2014).
- Ouverney, C. C., and J. A. Fuhrman, "Marine planktonic archaea take up amino acids." Appl. Environ. Microbiol. 66(11), 4829 (2000).
- Pester, M., C. Schleper, and M. Wagner, "The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology." Curr. Opin. Microbiol. 14(3), 300 (2011).
- Francis, C. A., J. M. Beman, and M. M. M. Kuypers, "New processes and players in the nitrogen cycle: The microbial ecology of anaerobic and archaeal ammonia oxidation." ISME J. 1(1), 19 (2007).
- Mussmann M., et al., "Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers." Proc. Natl. Acad. Sci. Unit. States Am. 108(40), 16771 (2011).
- Parada, A. E., D. M. Needham, and J. A. Fuhrman, "Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples." Environ. Microbiol. doi:10.1111/1462-2920.13023 (2015).
- Santoro, A. E., K. L. Casciotti, and C. A. Francis, "Activity, abundance and diversity of nitrifying archaea and bacteria in the central California current." Environ. Microbiol. 12(7), 1989 (2010).
- Pernthaler, A., J. Pernthaler, and R. Amann, "Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria." Appl. Environ. Microbiol. 68(6), 3094 (2002).
- Pernthaler, A., and J. Pernthaler, "Fluorescence in situ hybridization for the identification of environmental microbes." Meth. Mol. Biol. 353, 153 (2007).
- Teira, E., et al., "Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean." Appl. Environ. Microbiol. 70(7), 4411 (2004).
Publications and Presentations
- Dekas, A. E., Investigating climatically-relevant archaeal and bacterial metabolisms. (2013). LLNL-POST-639316.