Gabriela Loots (16-ERD-007)
Project Description
Traumatic injury renders an organism less tolerant to infection, with 40% of trauma patients dying from infection within about 76 hours of injury by unknown mechanisms. We plan to develop new approaches that will help us determine why. In recent years it has been hypothesized that a microbial gut–brain axis exists that is critical in modulating our immune system. More than 1,000 species of bacteria live within our gut in a commensal manner. These bacteria are vital to our health by supplying essential nutrients, synthesizing vitamins, aiding in digestion, influencing mood, and modulating nutrient and drug absorption. Commensal bacteria and probiotics also promote intestinal epithelial barrier integrity, and prevent antigens and pathogens from entering the mucosal tissues. Increasing evidence suggests that commensal bacteria contribute to our host defense by regulating metabolic equilibrium of the host immune system. We will generate and analyze data on biological molecules that translate into the structure, function, and dynamics of an organism and are derived from in vivo infection models that monitor genetic transcriptional expression in real time in both host and pathogen as a function of disease progression and traumatic injury. We will create a framework for generating in-depth genomic, molecular, and drug-related data that can be used to model the host-pathogen immunity gut–brain interactions in a whole organism. The long-term goal is to understand mechanisms of tolerance to pathogens, and to identify novel molecules that can be used therapeutically to reduce sepsis (toxic infection) in trauma patients.
We expect to demonstrate that both host and pathogen in an organism are dynamic and synergistically change and respond to each other. We aim to examine pathogen–host interactions at the "ecosystem level," meaning that we will systemically examine changes in both host and biotic community (biome) composition, gene expression, and protein interactions. In addition, we will view humans as a highly intricate, multi-directional communication network, where the gut, immune system, and the brain are constantly communicating with and adjusting to the needs of the organism. We intend to perform the first comprehensive biological study examining pathogen–host interaction at the organism's ecosystem level, and as a function of trauma. We will determine how traumatic injury affects the brain, the gut biome, and mucosal immunity. We will also determine how host tolerance to pathogens is affected by post-traumatic injury. Through proof-of-principle experiments in an endotoxin-induced lipopolysaccharide inflammation model of sepsis, we will identify molecular pathways that decrease tolerance to infection subsequent to trauma. Lipopolysaccharides are large molecules found in the outer membrane of certain bacteria, and elicit strong immune response in animals. We will also determine how drug metabolism and disposition, and gut biome are affected as a function of traumatic injury, inflammation, and exposure to antibiotics, because absorption in the gut modulates drug absorption.
Mission Relevance
Our research aims to take a radical approach to how we view human health in relationship to pathogen infections and commensal bacteria. By understanding pathogen–host interactions at the ecosystem level, systemically examining changes in both host and biome composition, gene expression, and protein interactions, we will support the Laboratory's core competency in bioscience and bioengineering. In addition, our methodology for meta-genomic analysis of biome samples is applicable to the core competency in high-performance computing, simulation, and data science.
FY16 Accomplishments and Results
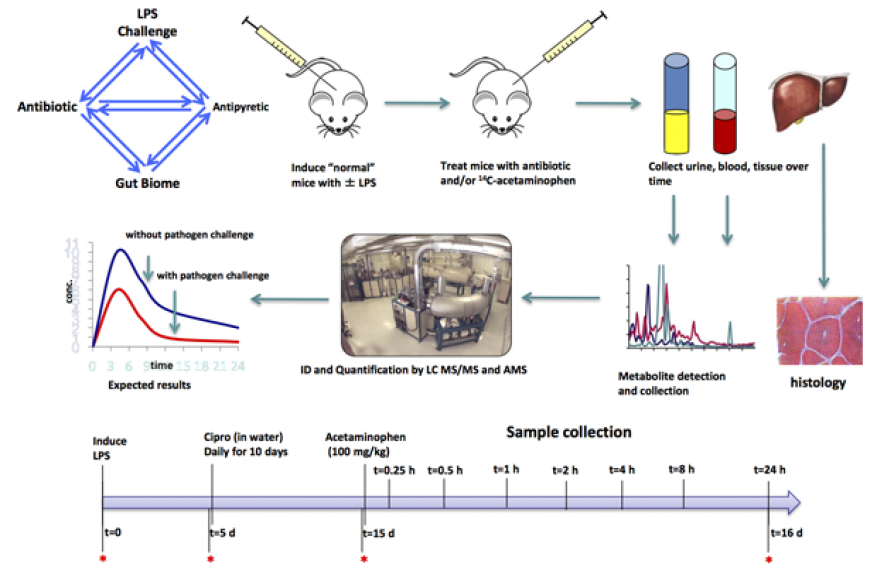
In FY16 we (1) bred cohorts of mice with genetically altered gut absorption; (2) initiated a lipopolysaccharide challenge of these mice; (3) challenged C57BL/6 mice (genetically modified mice commonly used as models of human disease) with a low dose of lipopolysaccharide and treated with the Cipro antibiotic; (4) determined how tylenol labeled with carbon-14 moves through mice as a function of challenge and antibiotic treatment; and (5) collected 128 samples of mouse blood, urine, and tissues. See figure below.
Publications and Presentations
- Malfatti, M. A., et al., Determining the role of the gut microbiome in altering drug metabolism. 2016. LLNL-POST-684459.