Sergio Wong (15-SI-002)
Project Description
The threat of a biological attack requires continued research into medical countermeasures. However, development of novel drug therapies is often hampered with costly failures in clinical trials because of a persistent inability to predict the useful or harmful effects of drugs. The heart is one major organ in which drug toxicity is particularly problematic and difficult to predict reliably. We aim to create and utilize a multiscale, human heart model using high-performance computing to predict the toxicity of drugs based on systems pharmacology. Our three-dimensional whole-heart model focuses on the prediction of drugs that are known to cause a prolongation in the heart beat, as well as on drugs that are known to be toxic for the heart. Results will be determined using clinically relevant, drug-induced perturbations to an electrocardiogram. We anticipate creating computational models for simulations at the cell, tissue, and organ scale.
We plan to develop a three-dimensional human heart model to demonstrate the usefulness of predicting drug effects on cardiac dynamics for drug design, preclinical screening, and predicting new therapeutics for specific arrhythmia syndromes. Our ultimate goal is to help shorten the drug development cycle and mitigate failures in expensive clinical trials. By integrating more-complex cellular mechanisms into Livermore's existing whole-heart model, including biological processes other than ion channel currents (the foundation of current cell-level models), as well as features of diseased hearts relevant to idiosyncratic toxicity, our computer model will accurately predict drug-induced cardiac toxicity and reveal biochemical mechanisms of toxicity that are manifested in electrophysiological abnormalities. Accurately predicting the pharmacokinetics of a drug candidate will dramatically reduce the time for the U.S. Food and Drug Administration approval of a new drug. This capability can be applied to all fields of pharmacology and toxicology.
Mission Relevance
The development of platforms and tools to reduce the time required to develop medical countermeasures for new pathogens by addressing key scientific barriers in the drug discovery and development process supports the strategic focus area in chemical and biological security. Our model to predict toxicity of new drug candidates in healthy and diseased hearts is also relevant to the Laboratory's core competency in bioscience and bioengineering.
FY16 Accomplishments and Results
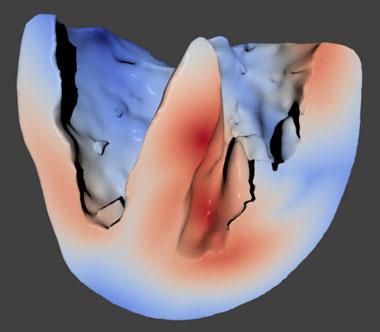
In FY16 we (1) tested and validated the newly completed whole-heart model (see figure); (2) incorporated additional ion-channel Markov-state models into the cellular model; (3) added a mitochondria model and biological consumers of adenosine triphosphate to the cardiac muscle cell model; (4) tested the more complex cellular model over various temperature ranges; (5) screened for tyrosine kinase inhibitor off-target human proteins using molecular modeling; (6) finalized the dissociation constant for ranolazine-binding hERG, the gene that codes for proteins forming the potassium ion channel in the heart and nervous tissue; (7) determined that the tyrosine kinase inhibitors decreased beating frequency in cardiac muscle cells; and (8) began simulations for clofilium- and chlorpromazine-binding hERG.