Crystal Jaing (14-ERD-081)
Abstract
We collaborated with Kansas State University in Manhattan, Kansas, to develop and evaluate two novel next-generation detection technologies to enhance biosecurity and to protect the agricultural economy. The first technology is a multiplexed assay to simultaneously detect ten swine viral and bacterial pathogens. The second is the Lawrence Livermore microbial detection array, which can detect more than 10,000 microbial species including 4,219 viruses, 5,367 bacteria, 265 fungi, 117 protozoa, and 293 archaea. We analyzed a series of swine clinical samples from past disease events to demonstrate the utility of the assays for faster and cheaper detection of emerging and foreign animal disease pathogens, and as a routine diagnosis and surveillance tool. A second goal of the study was to better understand mechanisms of African swine fever virus infection in pigs to aid the development of countermeasures and diagnostics. This virus has no vaccine. Although not currently in the U.S., African swine fever is on the rise on several European countries. A potential outbreak in the U.S. would be detrimental to the swine industry and the U.S. agricultural economy. We applied a genome-wide approach to characterize the pig immune responses after African swine fever virus infection. By using ribonucleic acid (RNA) sequencing and bioinformatics methods to identify genes and pathways that are affected during African swine fever infection, we identified a list of the most differentially expressed genes in the immune response pathways that could be used as candidate markers for disease outcome and diagnostics.
Background and Research Objectives
The identification and control of foreign animal diseases such as foot and mouth disease and swine fever are critical to the protection of U.S. agriculture and food systems. Past outbreaks have occurred in Europe, Central and South America, Asia, and Africa. The cost of eradicating a swine fever outbreak in the Netherlands in 1997 was about $2.3 billion. Swine fever is also a disease of concern for biological warfare. Early detection of disease outbreaks is crucial to the effective surveillance of emerging infectious diseases to safeguard animal and human health. Our goal was to develop new and effective surveillance technologies for early detection of known, emerging, and new foreign animal disease outbreaks; to enable biomarker discovery; and transition the technologies to national agricultural laboratories. In our collaboration with Kansas State University, which is the future location of National Bio and Agro-Defense Facility, we enhanced the scientific foundation for advanced research in agricultural and animal disease pathogens, as well as increased our engagement with the national agricultural pathogen surveillance efforts.
Our work involved developing at least two novel foreign animal disease and agricultural pathogen surveillance panels—a multiplexed Luminex bead assay to detect swine disease and near-neighbor species, as well as a broad-spectrum livestock pathogen-detection microarray. We collaborated with researchers at Kansas State University in agricultural pathogen surveillance and detection, and helped position the Laboratory to be more broadly engaged in developing techniques for detecting global animal disease outbreaks. Our goal was to transition both assays to Kansas State University as well as international collaborators. Additionally, we worked towards identifying genetic markers that could distinguish virulence capability for African and classical swine fever viral isolates as well as identifying potential diagnostic biomarkers for classical swine fever infection in pigs.
Scientific Approach and Accomplishments
Development of a Swine Respiratory Disease Multiplex Panel
We developed a swine respiratory disease multiplex panel to rapidly detect ten viral and bacterial pathogens:
- African swine fever
- Classical swine fever
- Porcine circovirus type 2
- Suid herpesvirus 1
- Porcine reproductive and respiratory syndrome
- Swine influenza
- Actinobacillus pleuropneumoniae
- Haemophilus parasuis
- Mycoplasma hyopneumoniae
- Streptococcus suis
The multiplex assay panel utilized Luminex-bead-based technology, which had several benefits. First, Luminex assays are higher throughput and more cost-effective than traditional polymerase chain reaction single-plexed assays. Second, most diagnostic labs have the instrument available and are already familiar with the technology. Third, Luminex beads are stable for up to 2 years at 4°C. Finally, the embedded African swine fever and classical swine fever beads can be removed for routine screening of domestic diseases. When there is an African swine fever or classical swine fever outbreak, or a need for increased surveillance of these fevers, the beads can be included into the panel for testing as deemed appropriate.
We performed sequence analysis of all available genomes from each of the pathogens and designed a set of deoxyribonucleic acid (DNA) signatures to detect the strains within each pathogen species. We performed experimental testing of various target and near-neighbor DNA samples and analyzed the sensitivity of the panel.1 The assay has been transitioned to Kansas State University for further evaluation with pig clinical samples. Additionally, a veterinary diagnostic company, Biovet, located in St-Hyacinthe, Québec, Canada, has also shown interest in licensing possibilities and has performed some evaluation of the assay panel.
Evaluation of the Lawrence Livermore Microbial Detection Array
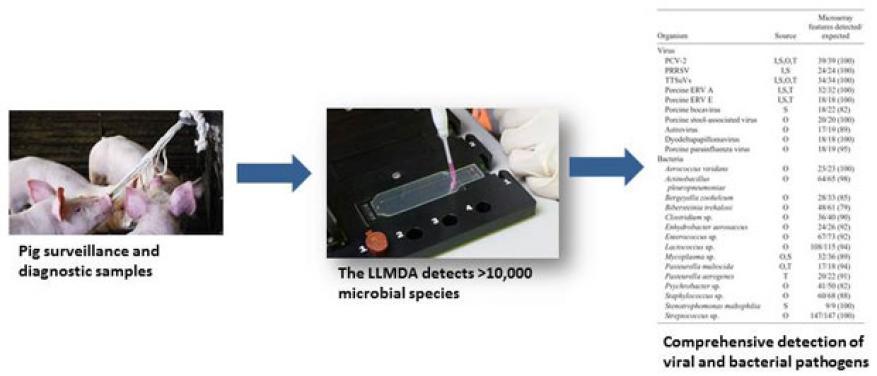
We also evaluated the Lawrence Livermore microbial detection array for use in detecting pathogens from swine clinical samples, and analyzed the array’s sensitivity and specificity (see Figure 1). This array is a comprehensive DNA detection technology capable of detecting any of the sequenced viruses, bacteria, fungi, and other microbial species. We tested clinical samples from a study using an experimental infection model for the analysis of porcine circovirus type 2 and porcine reproductive and respiratory syndrome, the two most common viral pathogens in the swine industry. The microbial detection array sensitively detected both pathogens. Additionally, as shown in Figure 2, the array detected several bacterial and viral co-infections in a number of clinical samples, which demonstrated the utility of this LLNL-developed technology in routine clinical diagnosis and surveillance.2 We also conducted a follow-on study using the microbial detection array to assess the association between pig microbiomes and the health outcomes of experimentally infected pigs.3
The array has been transitioned to Kansas State University and is now in use as a detection and diagnostic tool for swine viral and bacterial infections and microbiome studies.
Characterizing the Infection Mechanisms of the African Swine Fever Virus
We characterized the African swine fever virus infection mechanisms by conducting a comprehensive whole transcriptome analysis to determine the pig genes and pathways that are affected by this virus. This study was a collaboration with the Australian Animal Health Laboratory in Geelong, Victoria, Australia, which performed the pig infection, viremia, and phenotypic characterizations. We performed the RNAseq analysis using next-generation sequencing technology, the Illumina NextSeq, and bioinformatics methods to determine the most differentially regulated genes after African swine fever virus infection.4 Additionally, we compared the mechanisms of pigs infected with two types of African swine fever virus—a low pathogenic strain OURT88/3 and a high pathogenic strain GRG2007/1. We identified a list of the top-most differentially regulated genes from immune response pathways including macrophage function, natural killer cell and T cell function, viral activation, and lymphocyte activation. These genes could be used as potential markers for viral infection and diagnostics and to inform future vaccine development.
Additional Accomplishments
As part of developing a strategic partnership with Kansas State University, we participated in the education and training of graduate students and young faculty. We also hosted and trained three graduate students and two young faculty members in LLNL array and sequencing methodologies and bioinformatics tools to facilitate technology transfer to the university. As part of this technology transition, we also provided training on the multiplex assay, DNA microarray protocols, and the bioinformatic software for microarray analysis. We have also engaged in discussions with Kansas State University about broad collaborations in biodetection and medical countermeasures.
Additionally, we established a national advisory group with leaders from the Department of Homeland Security, the U.S. Department of Agriculture, and the National Pork Board to seek their advice on the broad utility of our technologies. Additionally, in collaboration with Kansas State University, we have submitted research proposals to the Department of Agriculture, Department of Homeland Security, and the Pork Board to study disease mechanisms of foreign animal disease pathogens, and to develop faster and cheaper detection technologies.
Impact on Mission
A comprehensive suite of novel technology platforms to rapidly detect and characterize foreign animal disease pathogens will help protect the agricultural economy, food supply, public health, and biosecurity of the nation. This effort aligns with the Laboratory's strategic focus area in chemical and biological security, which aims to develop innovative systems and capabilities to rapidly detect and effectively respond to intentional use of chemical agents, pathogens, or natural outbreaks of pandemic diseases.
Conclusion
In conclusion, we achieved our research goals and successfully transferred our technology to Kansas State University while setting a strong foundation for long-term strategic partnership with the National Bio and Agro-Defense Facility as well as with other federal stakeholders. The partnership between LLNL and Kansas State University is at many levels, including scientific research, education and outreach, and future joint funding opportunities. Additionally, we expanded our international collaborations with the Australian Animal Health Laboratory and started discussions with several African countries about emerging animal and zoonotic disease diagnosis and surveillance. We will continue to pursue additional research opportunities in the U.S. and with international partners. The Lawrence Livermore microbial detection array was commercialized by Affymetrix in Santa Clara, California. Affymetrix develops and provides innovative technologies that enable multiplex and parallel analysis of biological systems.
References
- Carrillo, C., et al., "Simultaneous detection of swine respiratory pathogens using a multiplexed detection assay." In preparation.
- Jaing, C., et al., "Application of a pathogen microarray for the analysis of viruses and bacteria in clinical diagnostic samples from pigs." J. Vet. Diagn. Investig. 27, 313 (2015). LLNL-JRNL-659677. http://dx.doi.org/10.1177/1040638715578484
- Niederwerder, M. C., et al., "Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2)." Vet. Microbiol. 188, 1 (2016). LLNL-JRNL-679863. http://dx.doi.org/10.1016/j.vetmic.2016.03.008
- Jaing, C., et al., Gene expression analysis of whole blood RNA from pigs infected with low and high pathogenic African swine fever viruses. (2016). LLNL-PRES-701989.
Publications and Presentations
- Carrillo, C., et al., Simultaneous detection of swine respiratory pathogens using a multiplexed detection assay. (2016). LLNL-PRES-680851.
- Jaing, C. J., Application of a broad-spectrum microbial detection array for the analysis of pig pathogens. (2016). LLNL-POST-665063.
- Jaing, C., "Application of a pathogen microarray for the analysis of viruses and bacteria in clinical diagnostic samples from pigs." J. Vet. Diagn. Investig. 27, 313 (2015). LLNL-JRNL-659677. http://dx.doi.org/10.1177/1040638715578484
- Jaing, C., 2016. Rapid detection of emerging pathogens using the Lawrence Livermore microbial detection array. (2016). LLNL-PRES-681957.
- Jaing, C. J., The all-inclusive pathogen detection platform: Microbial detection microarray. (2016). LLNL-PRES-672940.
- Jaing, C., et al., Gene expression analysis of whole blood RNA from pigs infected with low and high pathogenic African swine fever viruses. (2016). LLNL-JRNL-726418.
- Niederwerder, M. C., et al., "Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2)." Vet. Microbiol. 188, 1 (2016). LLNL-JRNL-679863. http://dx.doi.org/10.1016/j.vetmic.2016.03.008