Priyadip Ray (15-ERD-053)
Abstract
Recent technological advances have enabled the collection of increasingly-massive datasets, bringing about new opportunities as well as considerable computational challenges. In healthcare, for example, effective leveraging of electronic medical records (EMR), large-scale, heterogeneous datasets comprising static and dynamic observations of patient demographic and physiological status, offers the potential to characterize and identify at-risk patient populations earlier and more accurately, saving lives and reducing hospital costs. Two major characteristics of these data (and indeed others relevant to Livermore’s broader mission) present considerable computational challenges: 1) the scale (or number of observations of the population of interest); and 2) the variety of possible data types present. In the case of EMR data, patient observations may be static or dynamic, as well as real-valued, integer-valued or categorical. Few joint statistical models exist that can simultaneously accommodate multiple, heterogeneous data types and facilitate estimation in a scalable manner. The goal of this project is to develop new statistical approaches to jointly model large-scale heterogeneous datasets and to derive timely insights from these data via development of distributed model estimation routines. In this project, we extended and enhanced Livermore’s SparkPlug distributed statistical modeling framework to enable parallelized Bayesian inference, which led to the development of novel statistical models for the joint analysis of static and dynamic data types. These developments resulted in the demonstration of our scalable estimation approach on a large-scale EMR dataset provided by our collaborators at Kaiser Permanente Northern California (KPNC). In addition, this project produced numerous academic contributions including publication of one research article in the Journal of Biomedical Informatics and two research articles under preparation, as well as presentation of one talk and four poster abstracts at high quality international conferences for clinical informatics research. In addition to cultivating a fruitful relationship with our primary collaborator, KPNC, this project directly led to engagement efforts with other stakeholders in the healthcare community (e.g. University of Virginia Medical Center). In addition, this project supported the recruitment and training of new talent to the Laboratory, including two summer students, a post-doc, and a staff scientist. Overall, this research has helped broaden and enhance Livermore’s growing leadership in biomedical research.
Background and Research Objectives
The overarching goal of this project was to develop a new methodology and technical capabilities for the analysis of large-scale and complex multi-typed datasets, with our primary application being the retrospective modeling of nearly 250 thousand KPNC emergency department hospitalizations of suspected septic patients over a four-year period. Sepsis is a dysregulated immune response to infection that ranks as the leading and most expensive cause of in-hospital death in the United States (Liu et al. 2014). Early detection of the condition is vital to successful treatment outcomes (Liu et al. 2017). However, physiological heterogeneity of the condition from patient to patient as well as subtle changes undergone by a patient over time preclude timely detection. More generally, several characteristics of EMR data present technical challenges relevant to ongoing and future research initiatives at Livermore:
- their scale (incurring sample sizes in the hundreds of thousands to millions)
- their inclusion of multiple data types (e.g. static and dynamic features, integer- and real-valued, etc.)
- their tendency for missing entries
For this project, our objectives were to
- develop a scalable framework (built on Livermore’s SparkPlug platform) for distributed statistical inference and prediction in large-scale, complex datasets;
- characterize the clinically relevant and latent phenotypes in the KPNC EMR dataset by development and application of composite mixture models (CMMs); and
- create novel statistical models for the joint analysis of conventional clinical features as well as previously untapped data sources including administered medications and full trajectories of patient vital signs.
All objectives were achieved at the conclusion of this project, resulting in numerous presentations at international conferences and publication in a high-impact clinical informatics journal. We describe our specific accomplishments over the course of this project in more detail in the following sections.
Scientific Approach and Accomplishments
Livermore’s SparkPlug framework enables distributed (data-parallel) estimation of a wide range of statistical models, extending the powerful and popular Apache Spark distributed computing platform. Before this project, the SparkPlug framework did not provide support for Bayesian statistical inference (Markov chain Monte Carlo or MCMC methods) that can provide benefits including incorporation of prior knowledge in model development, quantification of uncertainty in model parameter estimates, and prediction of missing data entries accounting for this uncertainty. Recent years have seen the development of multiple successful approaches to parallelize MCMC-based inferences (Scott et al. 2016; Willie et al. 2013; Minsker et al. 2014) in which large-scale datasets are partitioned that are then distributed across nodes or cores in a parallel computing environment. MCMC runs are then carried out on each partition of the data with the resulting samples of the posterior distribution on model parameters pooled and processed to produce a posterior summary for the whole dataset. A significant achievement of this project was the extension of the SparkPlug framework to include a full suite of common prior distributions, implementations of multiple approaches to pool posterior inferences, and software utilities for large-scale data ingest and processing.
An important technical challenge this project overcame during this work was addressing the multi-typed nature of the patient feature vectors in the EMR dataset. For example, patient episode observations contained, among other features, integer-valued ages, categorical indicators of the facility where the patient had been treated, and real-valued summary statistics of the patient’s vital signs over the course of their hospitalization. To identify subgroups of patient episodes enriched for mortality events, we could not directly apply standard clustering techniques (Schlattmann 2009). To jointly model these multi-typed feature vectors, we leveraged composite mixture models (CMMs), a statistical model previously developed at Livermore (Wasson 2014; Sales et al. 2013) for developing scalable routines for the estimation of model parameters. The two central premises of the CMM are (1) that the dataset is assumed to be a mixture of different subpopulations or clusters (with potentially distinct characteristics and responses to interventions) and (2) that, given an indicator of the cluster to which the patient episode belongs, the dimensions of the feature vector are assumed to be independent of one another and modeled by univariate distributions appropriate for each dimension’s data type. For example, a feature dimension containing integer-count data might be modeled with a Poisson (rate of occurrence) distribution while another real-valued dimension representing the median systolic blood pressure of the patient over some post-admission period of their hospitalization might be modeled with a Gaussian (normal) distribution. We successfully demonstrated the efficacy of the CMM on a large-scale, multi-typed dataset designed to emulate the KPNC EMR dataset and found that we could estimate the true parameters of the model used to generate the simulated dataset with the same accuracy and in less time than carrying out a single MCMC run on a single node or core. We further demonstrated our scalable estimation routines by fitting a CMM to the KPNC EMR data, generating novel clinical insights about the physiological and demographic characteristics of the suspected septic patients.
In addition to our development efforts, two early statistical analyses of the KPNC EMR data were recognized at the 2016 International Conference of the American Thoracic Society (ATS). In the first analysis, we set out to benchmark mortality prediction performance on the KPNC EMR dataset using scalable implementations of two common classifiers (logistic regression and random forests). We found that a composite measure of the patient’s acute illness burden at admission to the emergency department was the most predictive feature of patient mortality by the end of hospitalization. Furthermore, we identified temporal trends in the features associated with patient mortality risk: physiological variables such as systolic blood pressure and respiratory rate averaged over longer post-admission periods became more relevant for mortality prediction than features available solely at the time of admission (e.g. the composite measure of acute illness). This analysis also uncovered novel co-clustering patterns among patient episodes that ended in mortality using the t-embedded stochastic neighbor embedding (t-SNE) approach (Maaten and Hinton 2017); regardless of clear, gender-based differences in feature space, patient episodes that ended in mortality tended to be embedded very close to one another. In the second analysis (Sales et al. 2016), another important outcome for this project, we enhanced mortality prediction in the KPNC sepsis dataset. Sepsis is typified by considerable physiological heterogeneity (Marshall 2014), suggesting that a patient population is likely composed of multiple subgroups. Thus, performance of a classifier fit to all patient features might suffer from averaging over these subgroups in which potentially different features might be predictive of mortality. By fitting classifiers to subpopulations of the patient episodes stratified by chronic and acute illness burden, we found that we could increase our sensitivity and identify salient features specific to the patient subgroups.
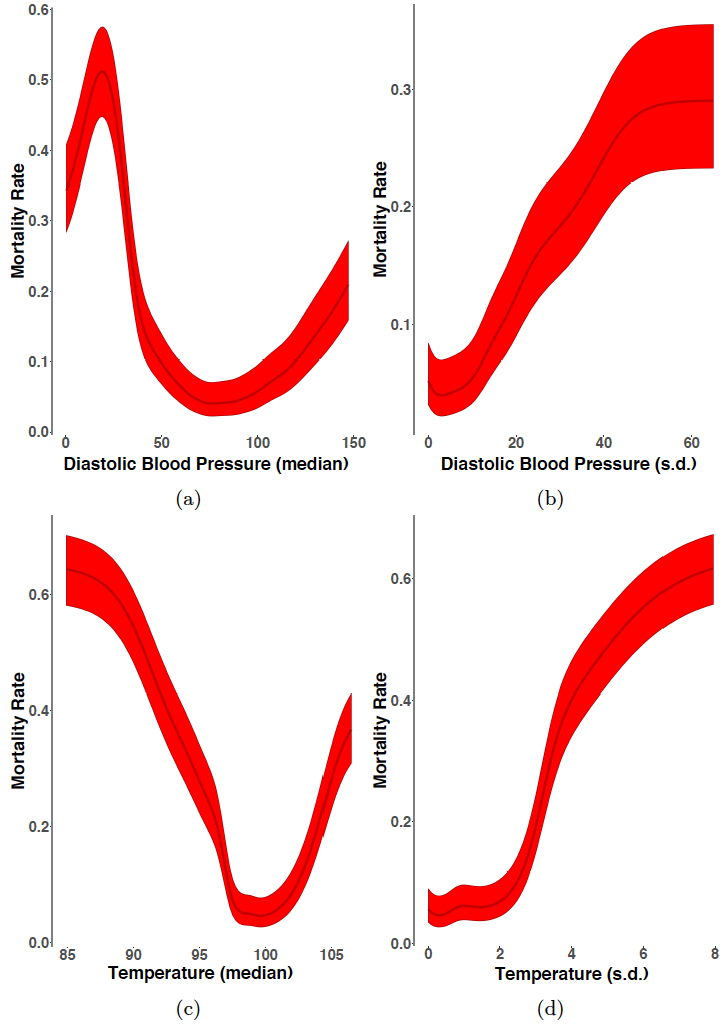
Another significant achievement of this project involved the development of stand-alone software packages for CMM fitting to multi-typed datasets. The SparkPlug extensions we previously developed enabled fast and accurate fitting of CMMs to large-scale multi-typed datasets. However, partly owing to idiosyncrasies of SparkPlug’s native programming language (Scala) as well as the absence of mature plotting utilities, our SparkPlug implementation of CMM fitting was cumbersome to use. We implemented the composite R software package, adapting the distributed expectation-maximization (EM) algorithm (Nowak 2003) to support fast, multi-core estimation of CMMs. In addition, we developed novel CMM-based plotting utilities; we adapted and enhanced a graphical technique used in conjunction with random forest classifiers (marginal importance plots) to CMMs, charting effects of changes (and uncertainty in those effects with confidence intervals) in individual physiological features on a patient’s risk of mortality during hospitalization.
In our CMM work, the primary features of interest were patient-centric: demographic and physiological information at and following the time of patient admission to the emergency department. However, EMR datasets (the KPNC dataset included) also tend to contain observations of the medications and procedures ordered and administered to the patient over the course of their hospitalization, giving valuable insight into the clinician’s intuition. Integration of these data (and development of best practices for their inclusion) into clinical informatics pipelines is still a nascent area of research, with the potential benefit of identifying the timing and type of effective interventions for patient treatment. As an important first step towards integration of medication information into our analyses, we developed a medication-wide association study (MWAS) to identify classes of medications associated with higher and lower risk of mortality during hospitalization. Our statistical analysis (Sales et al. 2017) identified 35 and 29 pharmacological classes associated with decreased and elevated risk of mortality, respectively, indicating adjunctive medications to which suspected septic patients might be responsive.
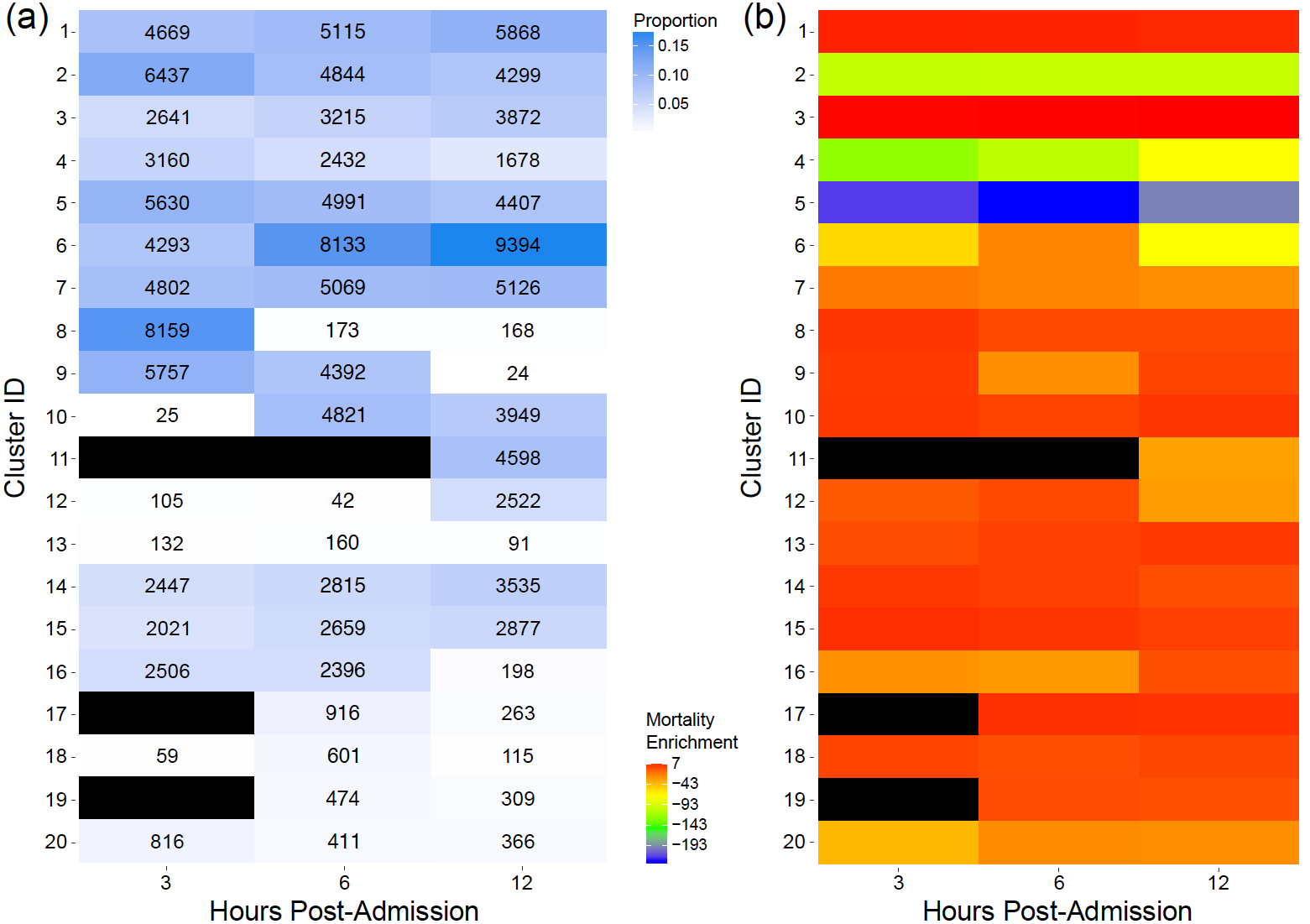
To understand the heterogeneity inherent in the suspected sepsis population and address the multi-typed nature of the patient observations, we developed and applied CMMs to the KPNC EMR dataset. However, this analysis faced two important practical challenges: 1) without clinically meaningful annotations (e.g. cluster 1 – higher mortality risk), the patient clusters had limited utility and 2) septic patient physiology was known to change over time, and independently fitting our CMMs to observations from different post-admission periods would have precluded analysis of such temporal patterns. To address these challenges, we generated patient features (including demographic information and vital sign summary statistics) for the 3, 6, and 12 hour periods immediately following admission to the emergency department. We then fit a CMM to all observations across all time periods, assigning patient episodes from different post-admission periods to the same set of clusters. In this way, we characterized all distinct constellations of patient features in the dataset, regardless of time, allowing analysis of patients with similar characteristics at different times in their hospitalization to be grouped in the same clusters. Evaluation of cluster-based approaches in clinical informatics generally involves benchmarking the inferred clusters against ‘gold standard’ labels (usually set by the clinical community and often difficult to count), leading to supervised (and potentially biased) analysis of patient phenotypes. To give these clusters more clinical interpretability, we developed an unbiased approach to annotate the inferred clusters by assessing whether patient episodes in the clusters were statistically significantly enriched for mortality events, as illustrated in Figure 1. We also constructed CMM-based marginal importance plots that illustrate the expected mortality rate given a particular vital value (Figure 2). This approach was generalizable to other categorical clinical outcomes, and, as our clusters were not restricted to pre-specified definitions, we were able to uncover new and subtle latent phenotypes. Moreover, owing to the structure of our model, we demonstrated additional clinical utility with competitive performance on missing data imputation, a common clinical informatics task.
Another achievement of this project involved the development of non-parametric Bayesian clustering techniques for electronic health care data, based on static and dynamic patient features. Clinicians routinely perform risk stratification and make treatment decisions for patients based on electronic health recordings (EHR). However, such decisions are typically made based on pre-set thresholds on patient vital signs. While such approaches have provided limited success, clinicians typically do not jointly consider available static information regarding the patients, such as co-morbidity and age, as well as dynamic information, such as time correlation properties of the EHR signals. However, both static and dynamic patient information provide valuable information regarding the underlying physiological state of the patient. We developed a novel non-parametric Bayesian model for jointly clustering patients based on both static and dynamic features. Our approach is based on a kernel stick-breaking process (KSBP)-based clustering of patient time series, in which individual patient time series are modeled with Gaussian processes (Ray et al. 2017b). Patients belonging to the same cluster are likely to share similar static features and share the parameters of the Gaussian process, which implies that the patients demonstrate similar dynamic behavior. The number of patient clusters as well as the parameters of the Gaussian processes are inferred from the data using MCMC techniques. Our approach is capable of handling both irregularly sampled as well as non-aligned patient time series data. Our results indicate that the proposed model can uncover distinct, recognizable patient clusters with medical significance. It was generally observed that patient clusters with high mortality rates were often associated with time-series which displayed short-range time-correlation and high volatility (Ray et al. 2017a).
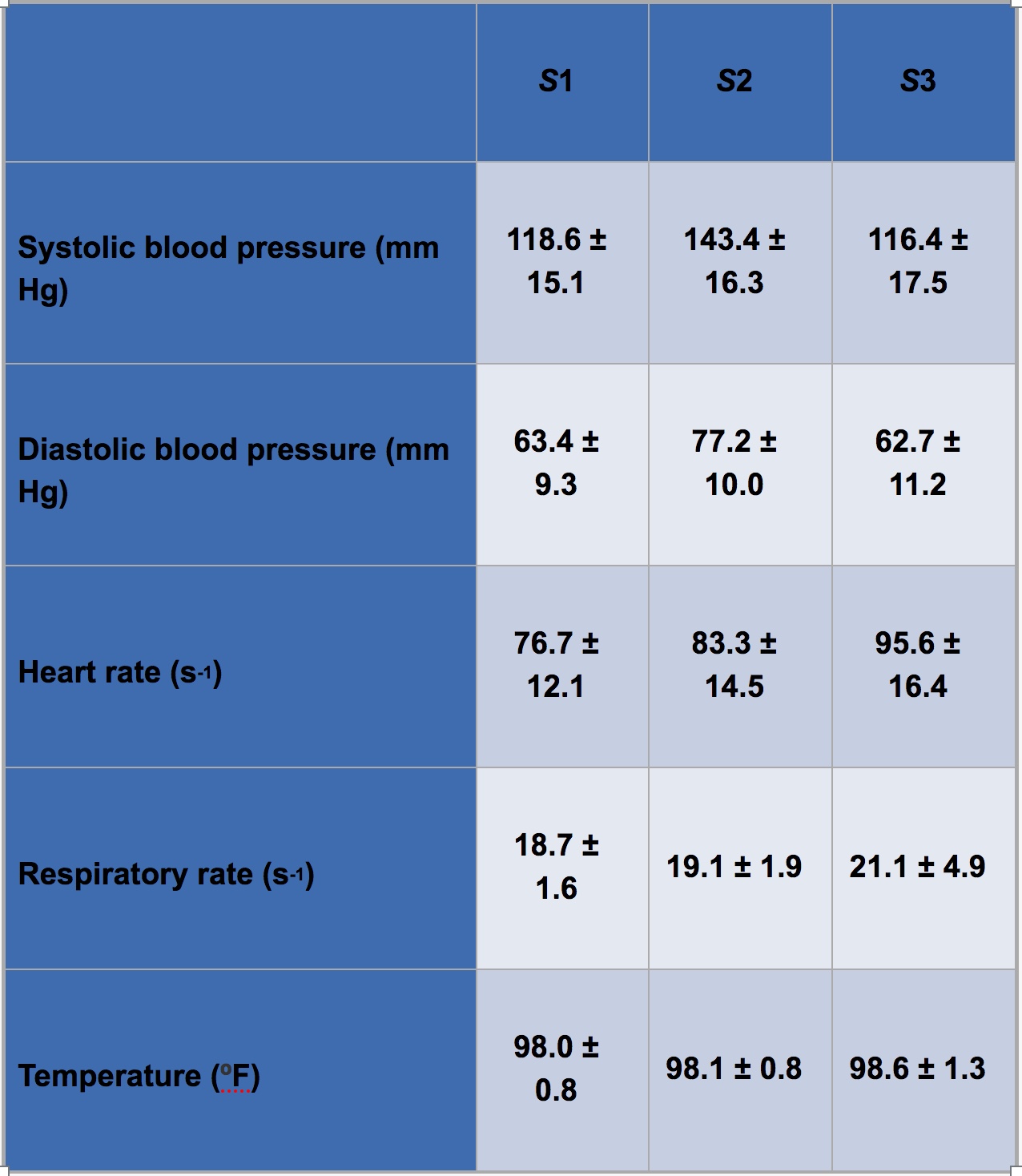
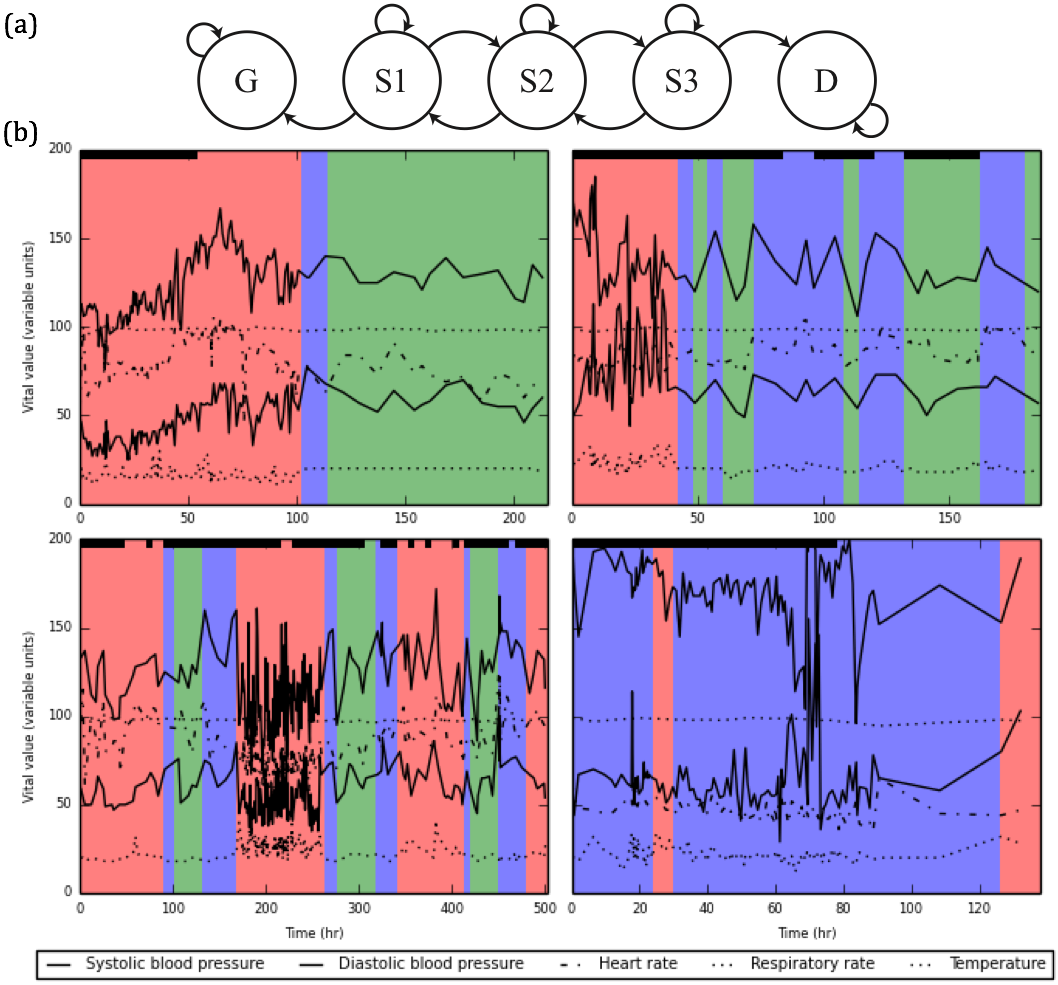
The final achievement of this project was the development of a discrete-time hidden Markov model to analyze a patient’s disease progression through sepsis. In this model, patients transition among three unobserved (latent) disease states (S1, S2, and S3) of increasing severity, ultimately ending in one of two outcomes: discharge or death (Figure 3A). Transition probabilities among adjacent states follow a proportional hazards model, in which nine global parameters determine baseline transition probabilities, and each patient’s covariates (age, acute disease burden score, and chronic disease burden score) are incorporated to provide patient-specific adjustments. Each discrete time step is associated with five vital observations (systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate, and temperature), which we model as conditionally independent (given a latent state) and normally distributed. Thus, the model captures both static and dynamic patient features. We performed Bayesian inference using MCMC techniques to infer (1) the nine global transition parameters, (2) mean and variance for each of the five vital observations from each latent state, and (3) the latent state of each patient at each discrete time step. Table 1 shows the inferred mean and standard deviation of the five vital observations from each latent state using a dataset of 20,000 patient hospitalizations. The inferred values from S3 (the most severe state) are consistent with clinical guidelines for sepsis diagnosis, providing a degree of qualitative validation to the model. Figure 3B shows characteristic disease trajectories of four patients. These plots provide an easily interpretable overview of a patient’s disease progression. The plots also demonstrate temporal smoothing in a patient’s disease trajectory compared to existing clinical guidelines, which typically rely on hard cutoffs to diagnose sepsis (Petersen et al. 2017).
Impact on Mission
This project has resulted in a suite of general methods, technical capabilities, and clinical informatics expertise that are immediately transferable to Livermore’s numerous research programs, particularly in the biomedical sciences. Our extensions to SparkPlug provide complementary tools for fast and accurate Bayesian statistical inference in a wide variety of models already or newly supported in the framework and can be applied in any setting involving complex, large-scale observations (e.g. cybersecurity efforts). The techniques we’ve developed for fitting and visualizing composite mixture models will enable immediate progress on other multi-typed datasets due to become available as part of collaborations with the Norwegian and American national cancer institutes, the San Francisco General Hospital Traumatic Brain Initiative, and the United States Veteran’s Administration, just to name a few. Our more recent methods developments for joint modeling of static and dynamic observations (HMM and KSBP) represent foundational technologies for the analysis of a wide range of complex, dynamical systems, especially those in which the researcher is interested in assessing the effects of interventions or perturbations from an external source (e.g. the Accelerated Therapeutic Opportunities in Medicine program).
This project has added to Livermore’s strong foundations for continued and impactful programs in biomedical informatics research and has led to the recruitment of full-time staff as well as two summer researchers. This project has also spawned valuable collaborations with clinical research teams at the University of Virginia and has provided a crucial platform for continued engagement with the biomedical research community via multiple aforementioned programs. Our team has represented Livermore at high-quality international conferences in critical care medicine and successfully published our analyses in a top-tier clinical informatics publication. This work has helped extend and enhance Livermore’s growing footprint and expertise at the high-impact intersection of machine learning and clinical research.
Conclusion
This project developed and demonstrated several major new capabilities. First, we extended and enhanced Livermore’s SparkPlug distributed statistical framework to enable parallelized Bayesian inference; second, we obtained clinically relevant and latent phenotypes in the KPNC EMR dataset via the application of composite mixture models (CMMs); finally we developed novel statistical models for the joint analysis of static patient features (such as age, acute disease burden score and chronic disease burden score) as well as dynamic patient features (such as full trajectories of vital signs). These capabilities lay the foundation for further research involving complex, large-scale heterogeneous observations as well as high-dimensional time-series observations, particularly in biomedical sciences, and cybersecurity.
References
Liu, Vincent, et al. 2014. "Hospital Deaths in Patients With Sepsis From 2 Independent Cohorts." JAMA 312 (1):90-92. doi: 10.1001/jama.2014.5804.
——— et al. 2017. "The Timing of Early Antibiotics and Hospital Mortality in Sepsis." American Journal of Respiratory and Critical Care Medicine 196 (7):856-863. doi: 10.1164/rccm.201609-1848OC.
Maaten, Laurens van der, and Geoffrey Hinton. 2017. "Visualizing Data using t-SNE." Journal of Machine Learning Research 9 (Nov):2579-2605.
Marshall, John C. 2014. "Why have clinical trials in sepsis failed?" Trends in Molecular Medicine 20 (4):195-203.
Mayhew, Michael B., et al. 2016a. "Evaluating Probabilistic Models and Scalable Inference for Large Scale Electronic Medical Records." Methodology for Precision Medicine Workshop, Durham, NC.
——— 2016b. "Identifying Time-Dependent Mortality Signatures in Cases of Suspected Infection Using Scalable Predictive Models." In A104. Critical Care: Sepsis Translational Insights, A2714-A2714.
——— 2017. "Flexible, Cluster-Based Analysis of the Electronic Medical Record of Sepsis with Composite Mixture Models." Journal of Biomedical Informatics (Accepted). Minsker, Stanislav, et al. 2014. "Scalable and robust Bayesian inference via the median posterior." Proceedings of the 31st International Conference on International Conference on Machine Learning - Volume 32, Beijing, China.
Nowak, Robert D. 2003. "Distributed EM algorithms for density estimation and clustering in sensor networks." IEEE transactions on signal processing 51 (8):2245-2253.
Petersen, Brenden K., et al. 2017. "Discrete-time hidden Markov model (HMM) for joint analysis of static and dynamic patient features." Under preparation.
Ray, Priyadip, et al. 2017a. "A Bayesian non-parametric framework for fusion of static and dynamic patient features." Under preparation.
——— 2017b. "Non-parametric Bayesian clustering of electronic health care data based on static and dynamic patient information." The 21st Annual Signal & Image Sciences Workshop, Livermore, California.
Sales, A.P., et al. 2013. "Semi-supervised classification of texts using particle learning for probabilistic automata." In Bayesian theory and applications. Oxford : Oxford University Press, 2013.
——— 2016. "Modeling Patient Subpopulations Improves Sepsis Mortality Prediction." In C95. Outstanding Epidemiology and Health Services Research in Critical Care, A6149-A6149.
——— 2017. "Medication-Wide Association Study in Sepsis and Suspected Infection." In A51. Critical Care: Risk Stratification and Prognostication - From Bedside to Big Data, A1816-A1816.
Schlattmann, Peter. 2009. "Medical applications of finite mixture models."
Scott, Steven L., et al. 2016. "Bayes and big data: the consensus Monte Carlo algorithm." International Journal of Management Science and Engineering Management 11 (2):78-88. doi: 10.1080/17509653.2016.1142191.
Wasson, T., and A. P. Sales. 2014. "Application-Agnostic Streaming Bayesian Inference via Apache Storm." Livermore-CONF-655453.
Willie, Neiswanger, et al. 2013. "Asymptotically exact, embarrassingly parallel MCMC." arXiv (preprint)1311.4780v2.
Publications and Presentations
Ray, Priyadip, et al. 2017. "Non-Parametric Bayesian Clustering of Electronic Health Care Data Dased on Static and Dynamic Patient information." The 21st Annual Signal & Image Sciences Workshop, Livermore, California. LLNL-POST-731002.
Sales, A.P., et al. 2016. "Modeling Patient Subpopulations Improves Sepsis Mortality Prediction." In C95. Outstanding Epidemiology and Health Services Research in Critical Care, A6149-A6149. LLNL-PRES-692041.
——— 2017. "Medication-Wide Association Study in Sepsis and Suspected Infection." In A51. Critical Care: Risk Stratification and Prognostication - From Bedside to Big Data, A1816-A1816. LLNL-POST-731326.