Jonathan Allen (17-ERD-062)
Abstract
Applying genome editing as a therapeutic tool is an important new disease-treatment strategy. To effectively design therapies that target individual genomes, it is important to have computational design tools that are informed by the entire complement of human genomic variation. Tools that search through populations of genes can identify off-target interactions that might cause a therapy to fail for an individual patient. This project explored the use of computational tools that differentiate small but potentially important differences between individual genetic variants with more accuracy than current tools, with the goal of developing genome-editing techniques that are designed to meet the needs of individual patients.
Background and Research Objectives
Ribonucleic acid (RNA) is a polymeric molecule that is (along with deoxyribonucleic acid, DNA) essential to the processes of coding, decoding, regulation, and expression of genes. Nucleic acids, lipids, proteins, and carbohydrates constitute the four major classes of molecules essential for all forms of life. Multicellular organisms use messenger RNA (mRNA) to convey genetic information. Guide RNAs (gRNAs) are the nucleic acids that guide the insertion and deletion of genetic material in mRNAs in a process known as genome editing. Editing events include insertion, deletion, and substitution of nucleotides within the edited RNA molecule. Designing an appropriate gRNA is an important element of genome editing. A gRNA can have unintended interactions (known as "off-targets") with other locations on the genome of interest. Designing gRNAs for precise genome editing remains a challenging problem due to the complex molecular mechanisms involved in genome modification, a phenomenon that is still not fully understood.
Computational tools (using rules derived in part from experimental observation and sequence similarity) are used to predict and avoid off-target effects and maximize the efficiency of genome editing. A fundamental limitation of existing tools is their inability to account for individual- and population-level genetic variability because they rely on the content of a canonical reference genome (i.e., a digital nucleic acid sequence database) and limited experiments to predict off-target interactions. An individual person’s genome differs from the reference genome by up to four million short variants (Jiang et al. 2015) with over 545 million variants identified and submitted to public databases. Tracking genomic loci (i.e., the fixed positions on a chromosome) with as many as four distinct variants relative to a candidate guide sequence could be important in influencing off-target interactions (Tycko et al. 2016). Taken together, these factors make it difficult to predict off-target effects in sub-populations, thus reducing the efficiency of genome-editing experiments and increasing the risk of unanticipated impacts on selected populations and individuals (Scott and Zhang, 2017).
With the continued development of whole-genome sequencing technology, it is feasible to access large amounts of individual human genomic data that can be used to track interactions of genome editing at the sub-population level. The objective of this project is to use the newly emerging gene-graph search tools to better understand the computational approaches for detecting changes in population-level genetic variation that could impact genome editing.
Scientific Approach and Accomplishments
The project used gene-graph search tools (for displaying genome-wide datasets) to evaluate the feasibility of accurately tracking permissive matches between reference genes in a population that targets a specific region of the genome for modification. Recently, Lawrence Livermore National Laboratory developed tools to support more sensitive and accurate searches of populations of genomes using a de Bruijn graph. These graphs are used in bioinformatics to assemble gene-sequencing reads for a genome. This tool was used to evaluate query sequences using a collection of reference genes that assist in gRNA design. Genome graphs have emerged as an important new tool for encoding individual genome variability in an efficient, searchable data structure (Paten et al. 2017; Eggertsson et al. 2017). Our research explored the possibility of using a de Bruijn population graph to differentiate genetic variants differing by one to five nucleotides, using raw genetic-sequencer reads as input, which would include gRNAs.
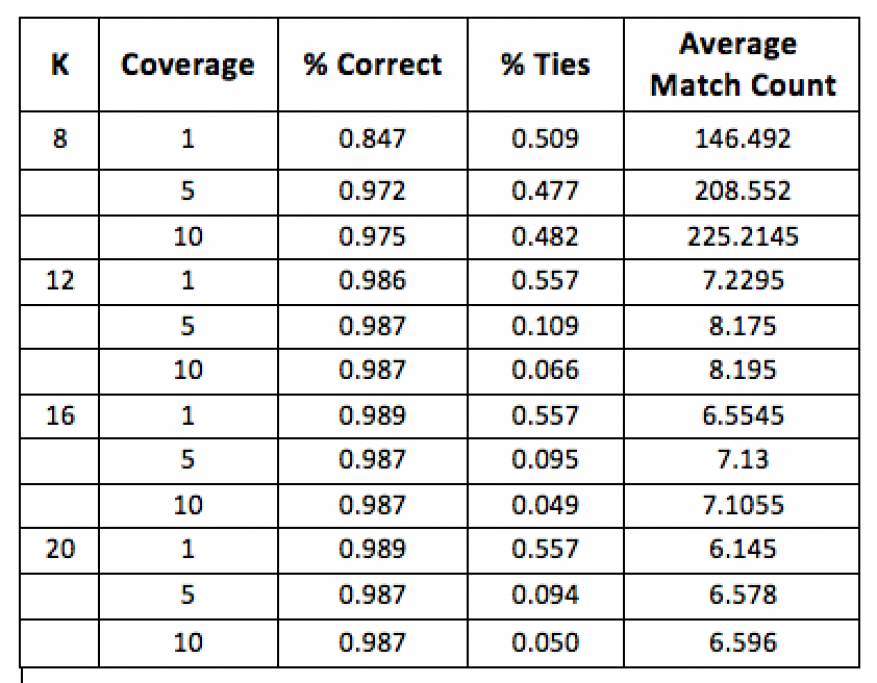
Gene-sequencer readouts are used as the query input cases where analysis is driven from a sequencing assay. In order to design a highly specific gRNA using sequence alignment to a reference-gene set, it is necessary to differentiate between the near-neighbor matches. In our work, each reference gene was serially replicated with point mutations five times; all variants were used to construct the population graph. Each gene was used as a query sequence after converting to a simulated short read form with simulated coverage of 1x, 5x, or 10x coverage. Accuracy was measured by the ability to retrieve the correct gene. The inference is that accurate gene differentiation between individuals will enable the design of gRNAs that can recognize small genetic differences introduced in individuals and sub-populations.
Table 1 shows cases where accurate target-gene retrieval is possible. The results indicate that a minimum starting exact match (k) of 12 is needed and that the gene must be covered at 10x coverage, otherwise the inability to disambiguate near matches (ties) increases from 0.066 to 0.109 percent.
Impact on Mission
This research supports the DOE and NNSA strategic goals of developing a strong biosecurity capability. Because genome editing is being developed as a new therapeutic tool for disease treatment, this research could open up new treatment options to respond to novel biological threats, providing the preliminary capability to develop new scalable computational tools to safely employ the use of genome-editing tools to enhance biosecurity, leveraging unique DOE computing resources.
Conclusion
The next steps for this work will be to extend collaborations with partners that maintain access to large collections of human genomic data that can be used for designing safe, patient-specific, genome-editing targets. In addition, it will be necessary to establish a collaboration with an experimental genome-editing group to test and validate the accuracy of new computational predictions. This will form the basis for developing a new computational capability that supports efficient and safe genome editing for clinical use.
References
Eggertsson, H.P., et al. 2017. "Graphtyper Enables Population-Scale Genotyping Using Pangenome Graphs." Nature Genetics 49, 1654−1660. doi: 10.1038/ng.3964.
Jiang, Y., et al. 2015. "The Missing Indels: An Estimate of Indel Variation in a Human Genome and Analysis of Factors that Impede Detection." Nucleic Acids Research 43, 7217–7228. doi: 10.1093/nar/gkv677.
Paten, B., et al. 2017. "Genome Graphs and the Evolution of Genome Inference." Genome Research. doi: 10.1101/gr.214155.116.
Scott, D.A. and F. Zhang. 2017. "Implications of Human Genetic Variation in CRISPR-based Therapeutic Genome Editing." Nature Medicine 23, 1095−1101. doi: 10.1038/nm.4377.
Molecular Cell 63, 355−370. doi: 10.1016/j.molcel.2016.07.004.