Felice Lightstone (17-ERD-043)
Executive Summary
We are exploring a novel, high-performance computational modeling method to design vaccines for highly mutable pathogens that may provide a groundbreaking solution to severe diseases such as human immunodeficiency virus and influenza. Our research could enable a universal flu vaccine to replace an annual one or rapidly design vaccines against natural pathogens or bioterrorism agents that are a threat to national security.
Project Description
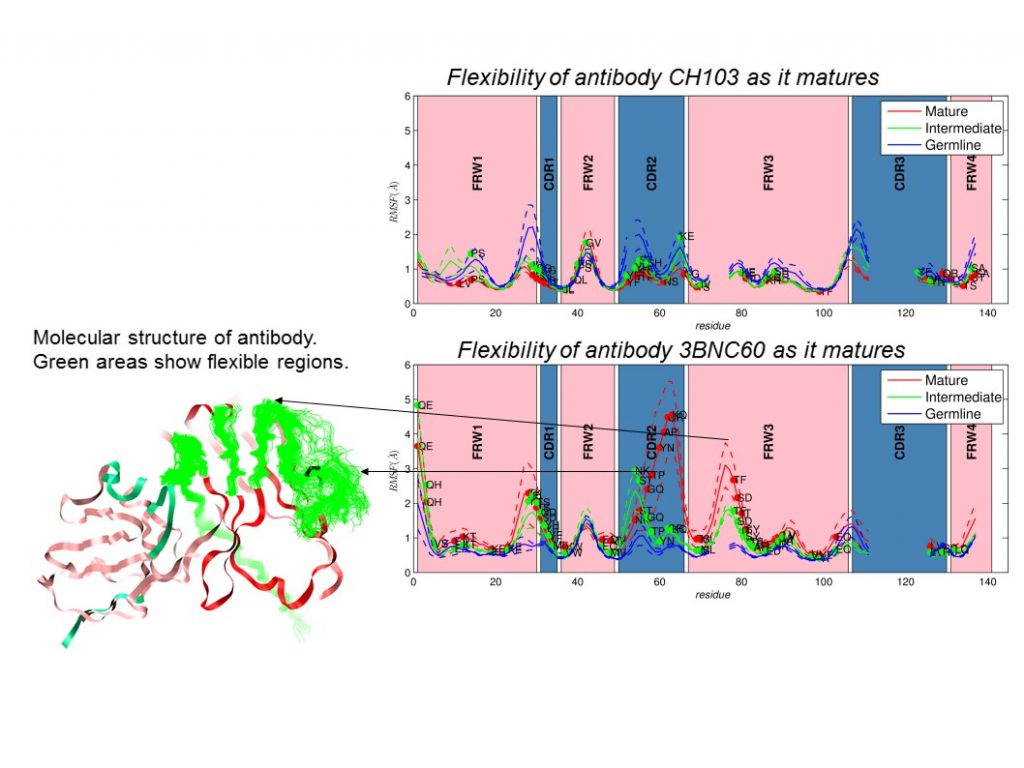
It has long been believed that vaccination will fail for highly mutable pathogens such as human immunodeficiency virus (HIV) and influenza. However, recent discovery of broadly neutralizing antibodies that can neutralize HIV by binding to highly conserved regions has revived interest in this area. Critical questions still remain regarding the identity, combination, and timing of antigens to develop broadly neutralizing antibodies into an immunization strategy. Potent antibodies are produced by a Darwinian evolution process, known as affinity maturation. Recently, it was demonstrated that inducing broadly neutralizing antibodies requires sequential immunization with multiple variant viral strains. We are designing these antibodies using a novel computational approach to provide vaccines for highly mutable pathogens. A holy grail in infectious disease is to rationally design vaccines capable of eliciting cross-reactivity for highly mutable pathogens that have defied vaccination so far. We intend to develop a novel multi-scale computational model of the affinity maturation process within a germinal center to predict the emergence of antibodies, using a spatially resolved, stochastic, agent-based model framework. Antigen internalization probabilities (in which antigen molecules are engulfed by a cell membrane and drawn into the cell) will be estimated from binding free energies calculated from all-atomic molecular dynamics simulations. HIV will serve as a case study, but the approach is readily extensible to influenza and other viral threats.
If successful, we expect to build a novel computational platform to design vaccines for highly mutable pathogens. It will provide a groundbreaking solution to combat severe diseases such as HIV and influenza and impact vaccine research by advancing development of a broadly cross-reactive vaccine against hemorrhagic fever viruses or a universal flu vaccine to replace an annual one. By developing a multi-scale model, we will address three specific aims: (1) develop a spatially resolved stochastic agent-based model that treats the key processes during affinity maturation accurately (using imaging experiments as a guide); (2) obtain the binding free energies of broadly neutralizing antibodies and antigen interactions from molecular dynamics simulations of atomistic models; and (3) develop a workflow to connect the agent-based model with the atomistic model and to automate simulations. Our research may produce the first high-fidelity, multi-scale computational model of affinity maturation that will be used to predict vaccination strategies for eliciting broadly neutralizing antibodies. Additional outcomes may be a mechanistic understanding and elucidation of the design rules that govern the universal emergence of these antibodies, as well as a significant first step in an effort to develop a predictive, computational model of adaptive immune response. Finally, our model framework will serve as an exemplar of coupling molecular dynamics of the protein domain involved in the biosynthesis of antibiotics and continuum modeling formalisms on a high-performance computing platform. We expect to enhance Lawrence Livermore's capabilities in bio-agent deterrence and medical research, particularly vaccine design, as well as to leverage leadership expertise at Harvard University and the Massachusetts Institute of Technology to build a new in-house, high-performance computing capability.
Mission Relevance
In support of Lawrence Livermore's chemical and biological security focus area, this research can provide a means to rationally and rapidly design vaccines against natural pathogens or bioterrorism agents that could be engineered to mimic their most virulent characteristics. Our research is also relevant to the DOE goal to deliver scientific discoveries and major scientific tools that transform our understanding of nature and strengthen the connection between advances in fundamental science and technology innovation.
FY17 Accomplishments and Results
In FY17 we worked to establish the protocols and methods to determine the necessary binding free energies of antibody and antigen interactions. Specifically, we (1) determined the minimal truncated size of the antibody and antigen complex that preserves accurate molecular dynamics; (2) determined that implicit solvation models for the antibody and antigen complex dynamics are adequate to use in molecular dynamics simulations; (3) established methods to generate these complexes using a computer program for protein structure calculation known as MODELLER; and (4) established a protocol using molecular mechanics energies combined with generalized Born and surface-area continuum solvation to calculate the protein-to-protein binding affinity.