Sahar El-Etr (15-ERD-017)
Abstract
We have developed a new approach to host-pathogen target discovery and verification that has wide applicability to the bio-security mission area. Using Burkholderia pseudomallei we utilized transcript capture and next generation sequencing (NGS) techniques to analyze gene expression in both pathogen and host over the course of infection in a human-host multicellular model. In parallel, we used multiple targeted proteomics approaches to identify host proteins produced by the pathogen during infection and to detect their potential interactions with host proteins. These data were used to build a multi-scale temporal model of infection. Using this model, we have identified 85 Burkholderia sp. extracellular, outer membrane or effector proteins that are predicted to interact with one or more human proteins. Of these proteins, 22 are predicted to interact with human immune-system proteins, potentially providing a novel hypothesis for how B. pseudomallei circumvents the host immune response. In addition, we have also identified 65 proteins (27 belonging to the immune system) that have been annotated as druggable targets. These proteins correspond to 337 FDA-approved drugs that could be tested as potential host-targeted therapeutics.
Background and Research Objectives
Burkholderia pseudomallei is a Gram-negative facultative intracellular bacterial pathogen that causes the disease melioidosis (Dance 2000). There are currently no licensed vaccines available for prophylaxis against the organism, and existing therapeutics lack efficacy (Bondi and Goldberg 2008). B. pseudomallei has been weaponized, possibly as recently as the early 1980s in Afghanistan, by the former Soviet military (Alibek 1999; Riedel 2004). Due to the weaponization history, its high potency in aerosol infection, and resistance to many common antibiotics, B. pseudomallei is considered a Tier 1 Select Agent (a biological agent that is highly threatening to public health) by the Centers for Disease Control and Prevention (CDC) (Dance 2000). The Defense Threat Reduction Agency’s Joint Science and Technology Office, Chemical and Biological Division is actively supporting research to develop novel medical countermeasures against B. pseudomallei. Research to identify therapeutic targets in both the pathogen and the host, which would enable development of effective anti-infective therapies and prophylactics (vaccines), is urgently needed (Bondi and Goldberg 2008). Of specific need is research that fully characterizes molecular mechanisms of B. pseudomallei pathogenesis in human-relevant model systems (Bondi and Goldberg 2008).
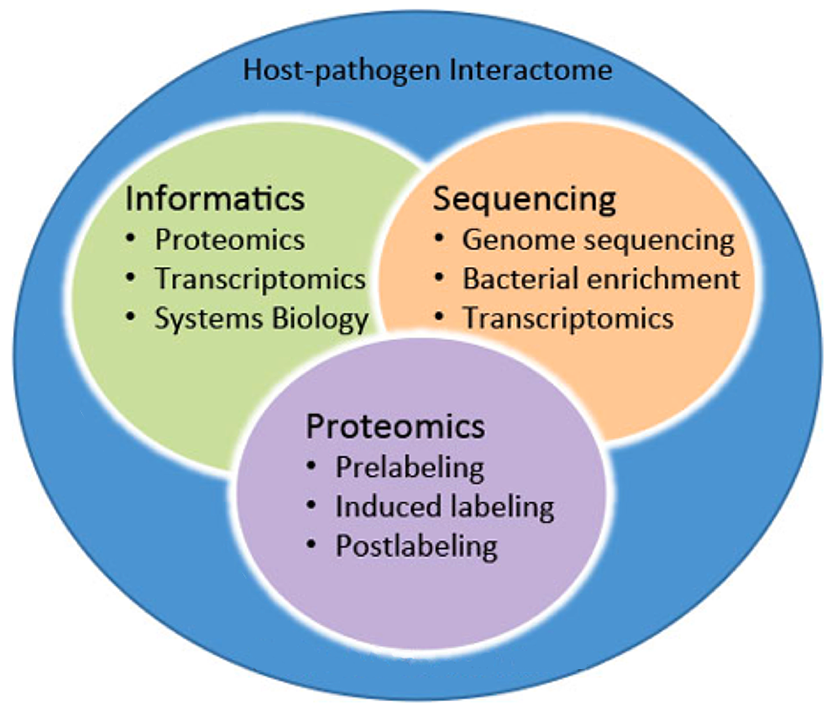
The goal of this project was to use a systems-biology approach (Figure 1) to identify and verify critical bacterial and host molecular mechanisms within the host-pathogen interactome that are involved in human infection by B. pseudomallei, thus establishing new targets for countermeasures against human melioidosis and a new strategy for target discovery that will be broadly applicable to other bacterial infections.
Recent technological advances now enable the comprehensive interrogation and modeling of molecular mechanisms of host-pathogen interactions at unprecedented sensitivity and throughput.
Scientific Approach and Accomplishments
We have demonstrated a new approach to target discovery and verification that uses cutting-edge technologies to surmount technical roadblocks that have confounded previous host-pathogen interaction studies. We developed a comprehensive, multi-scale temporal model of intracellular B. pseudomallei infection (Figure 2) using a systems-biology approach that leverages RNA-sequencing methods and technology established at Sandia National Labs (SNL) and focuses on B. pseudomallei clinical strains isolated from disease outbreaks in South East Asia.
Transcriptomics
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. The primary objective of the transcriptomics effort within this project was to generate global transcriptomic profiles of both B. pseudomallei ("Bp" in strain designations) and human host cells at different stages of infection for use in development of a comprehensive, multi-scale temporal model of host-pathogen interactions to elucidate the underlying molecular mechanisms and thereby identify new targets for anti-virulence countermeasures.
Accomplishing these objectives required five major technical advances: (1) modification of a key molecular-biology method for selective enrichment of pathogen-derived nucleic acids for successful use with Burkholderia sp. species; (2) NGS analysis of the genome features of a clinical isolate of interest (Bp PHLS6) to enable interpretation of transcriptomic profiles generated from this isolate; (3) establishment of two in vitro model systems for the study of B. pseudomallei infection; (4) NGS-enabled transcriptomic profiling analysis of both host and pathogen (i.e., dual RNA-sequencing) during B. pseudomallei infection; and (5) RNA-sequence analysis of the impact of amoebae-mediated priming on the pathogen transcriptome in order to identify the molecular mechanisms underlying the priming effect.
We started by sequencing the genome of strain Bp PHLS6. This strain was chosen for sequencing because it is a virulent clinical strain that has undergone minimal lab manipulation. It was originally isolated from a melioidosis patient in Bangladesh in 1960. The PHSL6 strain was also found to be highly efficient at entering and surviving inside amoebae, a mechanism thought to play a role in the prevalence of B. pseudomallei in soils (Moore 2010). The genome sequence was used for mapping reads derived from pathogen RNA-sequencing analysis of Bp PHLS6 infection cultures.
To establish our in vitro infection models, primary monocytes (i.e., a type of leukocyte, or white blood cell) collected from a single human donor were cultured under conditions that promoted their differentiation into macrophages (MDMs) and were then infected with B. thaitandensis, the lab strain Bp 1026b, or Bp PHLS6 at a multiplicity of infection (MOI) of approximately 20 to identify changes between fully virulent and mildly pathogenic strains, as well as potential differences between laboratory and clinical strains of B. pseudomallei. We found that the three B. pseudomallei strains were internalized by MDMs to similar degrees. These experiments were repeated with primary small-airway epithelial cells (SAEC); the results compared with those obtained with MDMs. Our results indicate that at an MOI of approximately 20, (1) Burkholderia sp. internalization is about 200 times more efficient in MDMs than in SAECs; (2) the intracellular Burkholderia sp. population within MDMs diminishes over time, presumably due to bacterial escape into the culture medium; (3) the intracellular B. thaitandensis population within the SAECs remains stable over time, presumably due to bacterial escape into neighboring cells; and (4) the intracellular B. pseudomallei population within SAECs grows over time, presumably due to bacterial escape into neighboring cells in combination with intracellular bacterial replication.
Through comparison of Burkholderia sp.-infected MDMs to mock-infected MDMs, we found that the Burkholderia sp.-infected MDMs differentially expressed hundreds of genes at each time point examined (3, 6, 12, 18, 24, and 36 hours post-challenge). Up-regulated (i.e., more highly expressed) genes outnumbered down-regulated genes throughout, the difference was initially vast (approximately 50 times more up-regulated at three hours post-challenge) then modest thereafter (5- to 25-times more up-regulated at six through 36 hours post-challenge).
Gene ontology (GO) term-enrichment analysis of the up-regulated genes indicated that they disproportionately belonged to pathways mediating pathogen recognition and cell-to-cell communication, which were activated immediately upon infection (i.e., by three hours post-challenge) and remained activated over the full time-course studied (i.e., to 36 hours post-challenge). Additionally, pathways mediating biosynthesis of and interaction with the extracellular matrix (ECM) were activated after an initial delay (i.e., by 6–12 hours post-challenge) and remained activated for the rest of the time course. Other pathways showing enrichment of up-regulated genes are not typically considered mediators of innate immune response to pathogens; it remains to be determined whether they do in fact participate in host response to B. thaitandensis. GO term-enrichment analysis of the down-regulated genes indicated that they disproportionately belonged to pathways mediating basic cellular functions that are typically repressed during macrophage activation (e.g., cell cycle, DNA replication). These pathways were repressed only at late stages of infection (24–36 hours post-challenge). There was also some evidence for transient repression of pathways involved in inflammation as well as late-stage repression of pathways not known to play a role in innate immunity.
Changes in gene expression are often driven by changes in the activity of transcription factors (TFs), which regulate sets of genes through recognition of cognate binding sites within the genes' promoters. In an attempt to identify the TFs responsible for observed changes in MDM gene expression during B. thaitandensis infection, we searched the promoter regions of the differentially expressed genes for conserved TF binding site (TFBS) sequence motifs. We found that throughout the time-course analyzed, many of the up-regulated genes had promoter regions that included TFBSs for the major constituents of the NF-κB (a protein complex that controls transcription of DNA) family and other major immune response regulons. The results support the idea that certain TFs (e.g., NF-κB) are consistently active throughout B. thaitandensis infection, whereas others (e.g., SRF) are active only at particular stages of infection. Consistent with the TFBS-enrichment results, we found that several TFs from the NF-κB family were induced throughout the infection time course. Additionally, two proteins primarily thought of as inhibitors of the NF-κB family (Nfkbia, Nfkbiz) were induced throughout the time-course as well.
Comparison of the host transcriptional responses to B. thaitandensis vs. Bp 1026b vs. Bp PHLS6 infection (through GO term-enrichment analysis of the up-regulated genes) indicated similarity in the activation of pathways mediating pathogen recognition, cell-to-cell communication, and biosynthesis of and interaction with the ECM. Pathways uniquely activated by B. thaitandensis infection are typical of host responses to other types of pathogens. Pathways activated by B. pseudomallei (but not B. thaitandensis) infection are involved in many different cell functions, most notably metabolic processes.
Surprisingly, we found that when using an MOI of ~20, SAECs showed virtually no significant transcriptional response to Burkholderia sp. infection, even using more lenient criteria for differential expression. Remarkably, this held true even in the late stages of B. pseudomallei infection (approximately 18 hours post-challenge), by which point the intracellular bacterial population had grown to roughly the same size as that initially observed in MDMs (three hours post-challenge). Moreover, we found that SAECs were fully capable of mounting a robust transcriptional response when challenged with B. thaitandensis at a higher MOI (2000, rather than 20, which resulted in internalization of about 10 times more bacteria).
Taken together, these results indicate that (1) MDMs are more sensitive in recognizing and/or responding to Burkholderia sp. challenge than are SAECs; and (2) SAECs do not recognize and/or respond to intracellular Burkholderia sp. populations. Regarding the latter effect, it is possible that SAECs simply lack a mechanism for recognition of and response to intracellular bacteria; or that Burkholderia sp. are exceptionally good at evading recognition or in disrupting responses. This key difference between MDM and SAEC response to Burkholderia sp. infection could have a major effect on the development of countermeasures and should be further characterized.
As with MDM transcriptional response to Burkholderia sp. infection, the SAEC transcriptional response to B. thaitandensis infection was dominated by gene induction, with up-regulated genes outnumbering down-regulated genes throughout the time-course by a greater degree at the initial time point (approximately 17-fold at three hours post-challenge) than at subsequent time points (4- to 8-fold at approximately six hours post-challenge). In total, 6,740 genes were differentially expressed at one or more time points; of these, 5,751 genes were up-regulated only, 989 genes were down-regulated only, and three genes were up-regulated at some post-challenge times and down-regulated at others.
Comparison of the genes differentially expressed by MDMs vs. SAECs in response to B. thaitandensis challenge revealed several interesting trends. Gene-induction programs elicited in both cell types upon B. thaitandensis infection are similar, whereas gene-repression programs are not. GO term-enrichment analysis of the genes up-regulated in SAECs indicated that several of the pathways that were activated in MDMs were also activated in SAECs, with roughly the same timing. However, no other pathways were robustly or consistently activated in SAECs; conspicuously inactive were pathways mediating pathogen recognition, cell-to-cell communication, and ECM biosynthesis and interactions. Analysis of genes down-regulated in SAECs revealed that the few pathways repressed in MDMs were similarly repressed in SAECs. Taken together, these results indicate modest overlap in the pathway activation profiles of MDMs versus SAECs challenged with B. thaitandensis, with an additional set of pathways activated only in MDMs.
In summary, transcriptional profiling of host response to Burkholderia sp. infection revealed that (1) MDMs respond robustly to challenge with small numbers of Burkholderia sp. (MOI 20), whereas SAECs respond robustly only to much larger numbers of bacteria (MOI 2000); (2) intracellular persistence and replication of Burkholderia sp. within SAECs does not provoke host response, indicating that SAECs do not sense the intracellular bacteria and/or their responses are suppressed by the bacteria; (3) for both MDMs and SAECs, host transcriptional responses tend to be dominated by up-regulation of genes and pathways; (4) MDMs respond to B. thaitandensis and B. pseudomallei in much the same way, such that pathogen recognition and signaling pathways are up-regulated initially, and those involved in cell–cell and cell–ECM interactions are up regulated after a delay. However, B. thaitandensis infection leads to activation of a set of pathways typically involved in responses to other types of pathogens; and B. pseudomallei infection leads to activation of metabolic pathways, which is a specific difference between pathogenic and nonpathogenic strains. In all cases, Bp 1026b and Bp PHLS6 provoked similar responses from MDMs; (5) SAECs respond to B. thaitandensis infection very differently as compared to MDMs; in particular, the pathogen recognition and signaling pathways are generally not activated and fewer pathways involved in cell–cell and cell–ECM interactions are activated in SAECs.
Proteomics
Though RNA-sequencing is a powerful tool delivering exquisite sensitivity, specificity, and quantitation over a large dynamic range, gene expression levels alone are insufficient to characterize the state of a cell system, since gene expression levels and the relative abundance of the encoded gene product (e.g., protein) are uncorrelated. While proteomic techniques have advanced significantly with sequencing technologies, proteomics studies of mixed cultures are still challenging as intracellular bacterial proteomes are generally obscured by the high amount of host proteins. Selective enrichment of bacterial proteins is therefore critical for mixed proteomic studies.
In this project, we employed a method previously established for the capture of the proteome of the intracellular pathogens Yersinia enterocolitica and Toxoplasma gondii (Mahdavi et al. 2014, Wier et al. 2015). This method employs bio-orthogonal noncanonical amino acid tagging that allows for specific labeling of bacterial proteins and the subsequent enrichment of bacterial proteome from co-culture conditions. The enrichment is achieved by expression of variant methionyl-tRNA synthetase (MetRS) in a bacterial genome, which alters the amino-acid sequence of proteins by incorporating azidonorleucine (Anl) in the place of wild-type methionine (Met). The azide tag on azidonorleucine then enables for enrichment of bacterial proteins and purification of proteins from virtually any growth condition. Anl is not utilized by the wild-type MetRS, so only the bacteria (and not host cells) incorporate the tag. This method was shown to have no significant effect on the growth and infectivity of other pathogens and represents a powerful tool to study the biology of intracellular pathogens.
To characterize the B. pseudomallei proteome, we expressed the variant Escherichia coli MetRS (MetRSNLL) in Bt and Bp ΔpurM via the insertion of a transposon (a sequence of DNA that is "transposable" within the genome of the cell). Our data show that expression of MetRS in Burkholderia sp. leads to labeling of newly synthesized bacterial proteins. Mass spectrometer analysis revealed that nearly 30 percent of methionines are replaced by azidonorleucine in labeled bacteria. Since on average there are approximately six methionines per bacteria, the estimated amount of label is 2 Anl per protein molecule. This labeling efficiency is enough to accomplish a proteome coverage that will be sufficient to perform quantitative proteomic studies.
We have also shown that this method can be employed to visualize intracellular Burkholderia sp. after host cell infection. Our recent experiments confirmed that the MetRS method enriches for Bp proteins replicating inside host cells. As expected, the overwhelming majority of peptides identified by MS analysis of whole cell lysates of infected host cells were human peptides. Out of approximately 13,000 unique peptides only 40 bacterial proteins were identified in the MS search and most of the remaining peptides were matched to the human genome. The majority of peptides detected in affinity-purified samples (where an Anl tag was used to enrich for bacterial proteins) were matched to bacterial proteins. These samples contained only low amounts of host protein. This data shows that our technique was successful for enrichment and characterization of the Burkholderia sp. proteome from intracellular bacteria. In addition, this tool will allow for the identification of secreted bacterial proteins that play an important role in bacterial virulence.
In parallel to the above method, we developed a protocol to post-label proteins with free N-terminal primary amines, by adapting a protocol previously reported for studying newly proteolyzed proteins in eukaryotic cells. Most eukaryotic proteins, or host protein, are N-terminally acetylated, where as most bacterial proteins have a free N-terminal primary amine (Polevoda and Sherman 2003). Using an engineered enzyme, subtiligase, which catalyzes the ligation of peptidic C-terminal esters to free N-terminal amines we can attach a synthetic peptide with an affinity tag to the N-terminus of proteins (Yoshihara, Mahrus et al. 2008). The affinity tag can then be used to enrich for tagged proteins, which can be subsequently analyzed by LC-MS/MS. This method complements the genetic engineering method using modified tRNA with ANL in that there is no genetic manipulation of the pathogen, making it applicable to any strain. The post-labeling strategy with subtiligase also enables the identification of newly proteolyzed host proteins, which are likely to play a role in the infection. We have demonstrated that subtiligase can be used to affinity purify bacterial proteins post-lysis and that there are differences between coculture lysates and controls (i.e., uninfected host-cells) as assessed by western blot (a technique used to detect specific protein molecules from among a mixture of proteins, in this case a total cell lysate).>
Systems Biology
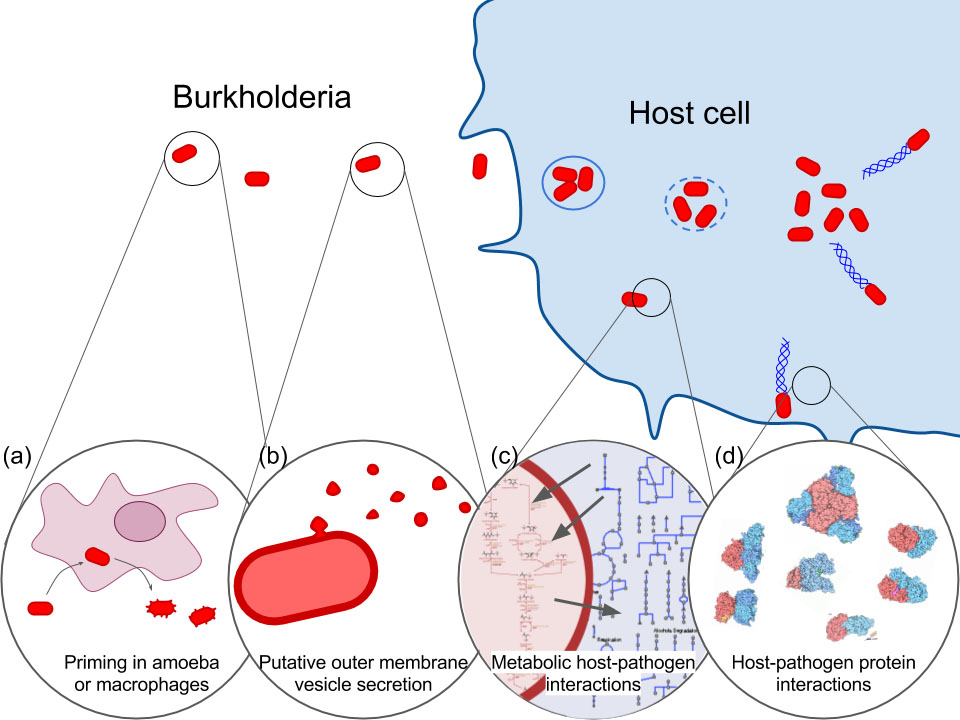
In order to study the systems biology of intracellular B. pseudomallei, we first had to characterize the systems biology of the pathogen and the host in isolation, and then investigate where and how these two systems interact. We focused specifically on two types of interactions between host and pathogen: direct host-pathogen protein-protein interactions, and metabolic interactions between the two organisms. These analyses were made possible by our genome sequencing efforts (Bp PHLS6), augmented and curated functional annotation, computational methods for prediction of protein–protein interactions, SNL RNAseq-enrichment technologies, and proteomics methods that we adapted for use in Burkholderia sp.
Like other Burkholderia sp. species, B. pseudomallei strains exhibit a remarkable level of genomic plasticity (Holden et al. 2004; Tumapa et al. 2008; Sim et al. 2008). The high abundance of simple sequence repeats (SSRs) in B. mallei and B. pseudomallei strains drives rapid genomic adaptation (Song et al. 2009) even during the course of acute infection in a single patient (Price et al. 2010) or short-term passaging in vitro (U'Ren et al. 2007). Due to the many repeats, genome sequencing of Bp PHLS6 required multiple sequencing runs and a collaboration between Lawrence Livermore National Laboratory, Sandia National Laboratories, and Los Alamos National Laboratory (D'Haeseleer et al. 2016). SNL sequence data consisted of 10,692,593 NextSeq 150PE short reads and 3,500,043 long fragments (Nextera Mate Pair preps), which assembled into 40 scaffolds with 220 contigs (overlapping sequence of DNA). Los Alamos National Laboratory sequence data consisted of an additional 15,757,418 MiSeq 250bp short insert (300 ± 70 bp) PE reads, producing 70 contigs. Because of the high number of repeats in the genome, it was decided to combine the Sandia National Laboratories long-insert data and Los Alamos National Laboratory short-insert data, and merge the resulting assembly with the prior Sandia and Los Alamos National Laboratory assemblies in parallel Phrap (DNA sequencing software) to generate the final improved draft assembly of 39 contigs totaling 7,322,181 Bp (% G+C 68.1) and 6,156 coding regions, including 118 genes associated with antibiotic and toxic compound resistance, as well as 16 genes associated with cellular invasion and intracellular resistance. As we have shown in other work, functional gene annotation is often highly incomplete (Griesemer 2017). To compensate, we first augmented the existing genome annotation for Bp PHLS6 using an in-house computational tool (Leung et al. 2016) that includes enzyme annotation by EFICAz2.5 (Kumar and Skolnick 2012) and transporter annotation with TransportDB’s Transporter Automatic Annotation Pipeline (Elbourne et al. 2017), and then homologizing with highly curated metabolic models for B. cenocepacia and B. multivorans (Bartell et al. 2014). This augmented functional annotation was then used to construct a high-quality genome-wide metabolic model using Pathway Tools (Karp et al. 2016), on which we can map the bacterial transcriptomics and proteomics data.
Host-pathogen protein–protein interactions can be computationally predicted based on homology with experimentally validated protein interaction pairs. These so-called interologs have been used successfully to map the host-pathogen interactome for several human pathogens, including Escherichia coli, Yersinia pestis, Mycobacterium tuberculosis and B. mallei (Memisevic et al. 2015; Krishnadev and Srinivasan 2011; Zhou et al. 2014). We used HPIDB (Ammari et al. 2016) to predict putative interactions between 430 Bp PHLS6 proteins, with 1,181 human host proteins. To narrow the range of likely true interactions, we assigned predicted subcellular localization to the pathogenic proteins using PsortB (Yu et al. 2010), resulting in 240 predicted extracellular or outer-membrane proteins. We also used EffectiveDB (Eichinger et al. 2016) to annotate 1,063 putative effector proteins in Bp PHLS6. Integrating this information, we find 85 predicted extracellular, outer-membrane, or effector proteins that are also predicted to interact with one or more human proteins. Interestingly, this set includes 22 pathogen proteins predicted to interact with human immune-system proteins, potentially providing novel hypotheses for how Burkholderia sp. disables or circumvents the host immune response.
In addition, out of the 1,181 human proteins predicted to interact with Bp PHLS6 pathogenic proteins, there are 65 (including 27 immune-system proteins) that have been annotated as druggable targets in a recent comprehensive map of molecular drug targets (Eichinger et al. 2016), corresponding to 337 FDA-approved drugs that could be tested as potential host-targeted therapeutics.
Using the pathogen-capture enrichment approach pioneered by Sandia National Laboratories (Bent et al. 2013), we were able to generate a transcriptomics time course for both the intracellular pathogen and the host cells, during the initial hours of infection. In order to validate which of the putative Burkholderia sp.-effector proteins are indeed secreted, we studied the secretome of cells in liquid culture, by mass spectrometry of the supernatant, versus the cell pellet itself. Surprisingly, the supernatant proteome was quite complex, with 11,621 spectra from 888 proteins detected by MS, compared to 10,832 spectra from 982 proteins from the cell pellet. This level of complexity in the supernatant could indicate a substantial amount of cell lysis. However, since some of the most overrepresented proteins in the supernatant are outer membrane proteins, an alternative explanation may be that B. pseudomallei secretes outer-membrane vesicles when grown in a liquid culture. This has not been previously documented for B. pseudomallei, but it is a recently described and widespread phenomenon that has been observed in a very wide range of bacterial pathogens, including B. cepacia (Allan et al. 2003; Galka et al. 2008, 2017). Outer-membrane vesicles are known to play an important role in virulence, but may also carry periplasmic and cytosolic proteins, which may explain the rich secretome observed in this case.
Impact on Mission
This project supports the broad NNSA national security mission area and the Lawrence Livermore National Laboratory bio-security mission area. The success of this project has allowed us to develop key credentials and capabilities toward achieving the Laboratory’s aim of becoming the go-to laboratory for providing end-to-end solutions for emerging bio-threats. We believe our work will significantly contribute to the pipeline developing specific countermeasures against B. pseudomallei, a CDC, HHS, and USDA Tier 1 Select Agent. Furthermore, the techniques developed during this project will enable accelerated identification of new targets for countermeasures against a wide array of infectious agents.
Conclusion
We have identified a number of potential bacterial and host targets that appear to play a role in the infection process. These targets need to be experimentally validated and then confirmed to play a role in vivo in models with human-like immunological responses to bacterial infection. Some of these targets appear to be specific to virulent Burkholderia sp. species, an area that is currently of particular interest to DTRA and the Biomedical Advanced Research and Development Authority (BARDA). We are currently writing multiple proposals to both of these agencies to extend these studies and augment the infectome model we build with additional bacterial strains. We have also recently submitted a DTRA grant proposal for research into repurposing FDA-approved drugs for prophylaxis or treatment of respiratory melioidosis.
References
Alibek, K. S. , and S. Handelman. 1999. "Biohazard: The Chilling True Story of the Largest Covert Biological Weapons Program in the World – Told from the Inside by the Man Who Ran It." New York: Random House.
Allan, N. D., et al. 2003. "Putative Virulence Factors are Released in Association with Membrane Vesicles from Burkholderia cepacia." Canadian Journal of Microbiology 49 (10): 613-624. doi: 10.1139/w03-078.
Ammari, M. G., et al. 2016. "HPIDB 2.0: A Curated Database for Host-Pathogen Interactions." Database (Oxford) 2016. doi: 10.1093/database/baw103.
Bartell, J. A., P. Yen, J. J. Varga, J. B. Goldberg, and J. A. Papin. 2014. "Comparative metabolic systems analysis of pathogenic Burkholderia." Journal of Bacteriology 196 (2): 210-226. doi: 10.1128/JB.00997-13.
Bent, Z. W., et al. 2013. "Use of a Capture-Based Pathogen Transcript Enrichment Strategy for RNA-Seq Analysis of the Francisella Tularensis LVS Transcriptome During Infection of Murine Macrophages." PLoS One 8 (10): e77834. doi: 10.1371/journal.pone.0077834.
Bondi, S. K., and J. B. Goldberg. 2008. "Strategies Toward Vaccines Against Burkholderia mallei and Burkholderia pseudomallei." Expert Review Vaccines 7 (9): 1357-1365. doi: 10.1586/14760584.7.9.1357.
D'Haeseleer, P., et al. 2016. "Genome Sequence of the Historical Clinical Isolate Burkholderia pseudomallei PHLS 6." Genome Announcements 4 (3). doi: 10.1128/genomeA.00649-16.
Dance, D. A. 2000. "Melioidosis as an Emerging Global Problem." Acta Tropica 74 (2-3): 115-119. doi: S0001-706X(99)00059-5 [pii].
Eichinger, V., et al. 2016. "EffectiveDB—Updates and Novel Features for a Better Annotation of Bacterial Secreted Proteins and Type III, IV, VI Secretion Systems." Nucleic Acids Research 44 (D1): D669-D674. doi: 10.1093/nar/gkv1269.
Elbourne, L. D. H.,et al. 2017. "TransportDB 2.0: A Database for Exploring Membrane Transporters in Sequenced Genomes from All Domains of Life." Nucleic Acids Research 45 (D1): D320-D324. doi: 10.1093/nar/gkw1068.
Galka, F., et al. 2008. "Proteomic Characterization of the Whole Secretome of Legionella pneumophila and Functional Analysis of Outer Membrane Vesicles." Infection and Immunity 76 (5): 1825-1836. doi: 10.1128/IAI.01396-07.
Griesemer, M., et al. 2017. "Combining Multiple Functional Annotation Tools Increases Completeness of Metabolic Annotation." bioRxiv beta 160887. doi: 10.1101/160887.
Holden, M. T., et al. 2004. "Genomic Plasticity of the Causative Agent of Melioidosis, Burkholderia pseudomallei." Proceedings of the National Academy of Sciences of the United States of America 101 (39): 14240-5. doi: 10.1073/pnas.0403302101.
Jan, A. T. 2017. "Outer Membrane Vesicles (OMVs) of Gram-Negative Bacteria: A Perspective Update." Frontiers of Microbiology 8: 1053. doi: 10.3389/fmicb.2017.01053.
Karp, P. D., et al. 2016. "Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology." Briefings in Bioinformatics 17 (5) :877-890. doi: 10.1093/bib/bbv079.
Kumar, N., and J. Skolnick. 2012. "EFICAz2.5: Application of a High-Precision Enzyme Function Predictor to 396 Proteomes." Bioinformatics 28 (20): 2687-8. doi: 10.1093/bioinformatics/bts510.
Leung, E., et al. 2016. "Protein Sequence Annotation Tool (PSAT): A Centralized Web-Based Meta-Server for High-Throughput Sequence Annotations." BMC Bioinformatics 17: 43. doi: 10.1186/s12859-016-0887-y.
Mahdavi, A., J., et al. 2014. "Identification of Secreted Bacterial Proteins by Noncanonical Amino Acid Tagging." Proceedings of the National Academy of Sciences of the United States of America 111 (1): 433-8. doi: 10.1073/pnas.1301740111.
Memisevic, V., et al. 2015. "Mining Host-Pathogen Protein Interactions to Characterize Burkholderia mallei Infectivity Mechanisms." PLoS Computational Biology 11 (3): e1004088. doi: 10.1371/journal.pcbi.1004088.
Moore, E. A. "Interactions of Burkholderia pseudomallei and Acanthamoeba castellanii and Their Effects on Virulence in Human Monocytes." (master's thesis, Brigham Young University, 2010), https://scholarsarchive.byu.edu/etd/2581/.
Oruganty, K., and N. Srinivasan. 2011. "Prediction of Protein-Protein Interactions Between Human Host and a Pathogen and its Application to Three Pathogenic Bacteria." International Journal of Biological Macromolecules 48 (4): 613-9. doi: 10.1016/j.ijbiomac.2011.01.030.
Polevoda, B., and F. Sherman. 2003. "N-Terminal Acetyltransferases and Sequence Requirements for N-Terminal Acetylation of Eukaryotic Proteins." Journal of Molecular Biology 325 (4): 595-622. doi: 10.1016/S0022-2836(02)01269-X.
Price, E. P., et al. 2010. "Within-Host Evolution of Burkholderia pseudomallei in Four Cases of Acute Melioidosis." PLoS Pathogens 6 (1): e1000725. doi: 10.1371/journal.ppat.1000725.
Riedel, S. 2004. "Biological Warfare and Bioterrorism: A Historical Review." Proceedings (Baylor University Medical Center) 17 (4): 400—406.
Sim, S. H., Yet al. 2008. "The Core and Accessory Genomes of Burkholderia pseudomallei: Implications for Human Melioidosis." PLoS Pathogens 4 (10): e1000178. doi: 10.1371/journal.ppat.1000178.
Song, H., et al. 2009. "Simple Sequence Repeat (SSR)-Based Gene Diversity in Burkholderia pseudomallei and Burkholderia mallei." Molecules and Cells 27 (2): 237-41. doi: 10.1007/s10059-009-0029-8.
Tumapa, S., et al. 2008. "Burkholderia pseudomallei Genome Plasticity Associated with Genomic Island Variation." BMC Genomics 9: 190. doi: 1471-2164-9-190.
U'Ren, J. M., et al. 2007. "Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping." BMC Microbiology 7: 23. doi: 10.1186/1471-2180-7-23.
Wier, G. M., et al. 2015. "New Method for the Orthogonal Labeling and Purification of Toxoplasma gondii Proteins While Inside the Host Cell." American Society for Microbiology MBio 6 (2): e01628. doi: 10.1128/mBio.01628-14.
Yoshihara, H. A., et al. 2008. "Tags for Labeling Protein N-Termini with Subtiligase for Proteomics." Bioorganic & Medicinal Chemistry Letters 18 (22): 6000-3. doi: 10.1016/j.bmcl.2008.08.044.
Yu, N. Y., et al. 2010. "PSORTb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes." Bioinformatics 26 (13):1608—1615. doi: 10.1093/bioinformatics/btq249.
Zhou, H., S. et al. 2014. "Stringent Homology-Based Prediction of H. sapiens-M. tuberculosis H37Rv Protein-Protein Interactions." Biology Direct 9:5. doi: 10.1186/1745-6150-9-5.
Publications and Presentations
D’haeseleer, P., et al. 2016. "Genome Sequence of the Historical Clinical Isolate Burkholderia pseudomallei PHLS 6." American Society for Microbiology Genome Announcements 4(3): e00649-16. doi: 10.1128/genomeA.00649-16.
Franco, M., et al. 2016. "A Novel Method for Proteomic Profiling of Burkholderia." American Society for Microbiology Microbe 2016. Boston, MA, 16—20 June 2016.