Gabriela Loots (16-ERD-007)
Executive Summary
A growing body of evidence suggests that the composition of a person's gut bacteria can indicate disease risk as well as affect how a person heals injuries, tolerates infections, or responds to drug treatments. In light of this, we are taking a radical approach to how we view human health by generating the first system’s biology account of immune-gut and brain responses to simultaneous injury and infection. We are examining pathogen-host interactions at the ‘ecosystem level’ by systemically examining changes in the host and biome composition, gene expression, and protein interactions.
Project Description
Traumatic injury renders an organism less tolerant to infection, with 40% of trauma patients dying from infection within about 76 hours of injury by unknown mechanisms. We plan to develop new approaches that will help us determine why. In recent years, it has been hypothesized that a microbial gut–brain axis exists that is critical in modulating our immune system. More than 1,000 species of bacteria live within our gut in a commensal manner. These bacteria are vital to our health by supplying essential nutrients, synthesizing vitamins, aiding in digestion, influencing mood, and modulating nutrient and drug absorption. Commensal bacteria and probiotics also promote intestinal epithelial barrier integrity, and prevent antigens and pathogens from entering the mucosal tissues. Increasing evidence suggests that commensal bacteria contribute to our host defense by regulating metabolic equilibrium of the host immune system. We will generate and analyze data on biological molecules that translate into the structure, function, and dynamics of an organism and are derived from in vivo infection models that monitor genetic transcriptional expression in real time in both host and pathogen as a function of disease progression and traumatic injury. We will create a framework for generating in-depth genomic, molecular, and drug-related data that can be used to model the host–pathogen immunity gut–brain interactions in a whole organism. The long-term goal is to understand mechanisms of tolerance to pathogens, and to identify novel molecules that can be used therapeutically to reduce sepsis (toxic infection) in trauma patients.
We expect to demonstrate that both host and pathogen in an organism are dynamic and synergistically change and respond to each other. We aim to examine pathogen–host interactions at the "ecosystem level," meaning that we will systemically examine changes in both host and biotic community (biome) composition, gene expression, and protein interactions. In addition, we will view humans as a highly intricate, multi-directional communication network, where the gut, immune system, and the brain are constantly communicating with and adjusting to the needs of the organism. We intend to perform the first comprehensive biological study examining pathogen–host interaction at the organism's ecosystem level, and as a function of trauma. We will determine how traumatic injury affects the brain, the gut biome, and mucosal immunity. We will also determine how host tolerance to pathogens is affected by traumatic injury and treatment. Through proof-of-principle experiments in an endotoxin-induced lipopolysaccharide inflammation model of sepsis, we will identify molecular pathways that decrease tolerance to infection after trauma. Lipopolysaccharides are large molecules found in the outer membrane of certain bacteria, and elicit strong immune response in animals. We will also determine how drug metabolism and disposition, and gut biome are affected as a function of traumatic injury, inflammation, and exposure to antibiotics, because absorption in the gut modulates drug absorption.
Mission Relevance
Our research aims to take a radical approach to how we view human health in relationship to pathogen infections and commensal bacteria. By understanding pathogen–host interactions at the ecosystem level, systemically examining changes in both host and biome composition, gene expression, and protein interactions, we will support the Laboratory's core competency in bioscience and bioengineering. Identifying novel molecules to treat toxic infections is relevant to DOE’s strategic objective to transform understanding of nature and strengthen the connection between advances in fundamental science and technology innovation. In addition, our methodology for meta-genomic analysis of biome samples is applicable to the core competency in high-performance computing, simulation, and data science.
FY17 Accomplishments and Results
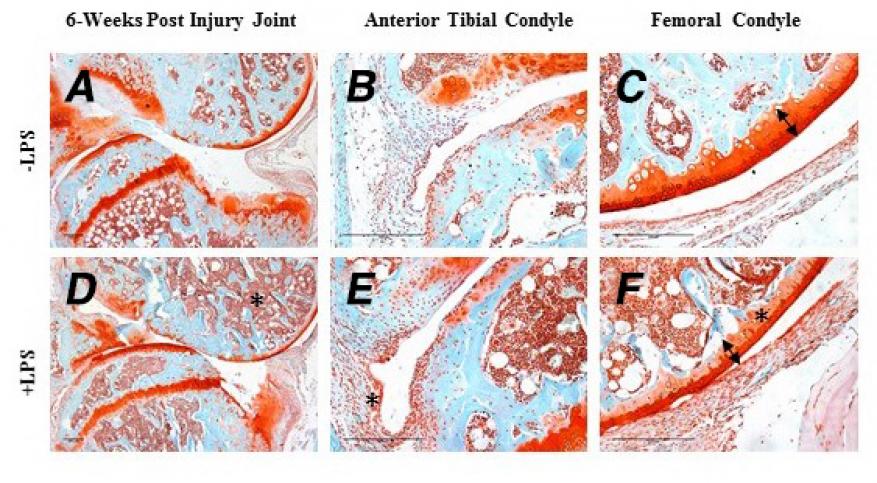
In FY17 we (1) treated four strains of mice (C57Bl/6, C3H/H3J, STR/ort, and Mrl/mpj), which have varying susceptibility to post-traumatic osteoarthritis, with antibiotics; (2) determined that antibiotic treatment in combination with injury induced 100% lethality in the C57Bl/6 strain of mice and partial lethality in the C3H/H3J strain mice 10 days after injury—lethality was prevented by administration of lipopolysaccharides in C57Bl/6 mice; (3) collected the joints of injured mice six weeks after injury (see figure); (4) collected the gut biome before antibiotic treatment, after lipopolysaccharides, and post-injury and examined using the LLNL microbial array; and (5) examined the gut histologically, and collected RNA for sequencing to determine the relationship between gut biome, immune system, and healing.
Publications and Presentations
Mendez, M. E., et al. 2017. "Effects of LPS Induced Inflammation Pre-Injury on ACL Rupture Mouse Models of Post-Traumatic Osteoarthritis." Orthopedic Research Society 2017 Annual Meeting, San Diego, CA, 19–22 March 2017. LLNL-ABS-737738.
Sebastian, A., and G. G. Loots. 2017. "Genetics of Sost/SOST in Sclerosteosis and van Buchem Disease Animal Models." Metabolism, October 25, 2017. Elsevier. 10.1016/j.metabol.2017.10.005. LLNL-JRNL-730352.