Mimi Yung (17-ERD-013)
Executive Summary
We are exploring a novel cell-based drug-delivery platform in which a therapeutic microbe is engineered to detect a specific pathogen and deliver antimicrobial protein fragments in high concentrations in the local pathogen environment. This research may have a broad impact on synthetic-biology solutions for biosecurity applications and antibiotic-resistant infections in both military and civilian healthcare environments.
Project Description
Antimicrobial peptides are strong therapeutic candidates for use against antibiotic-resistant pathogenic bacteria because they display rapid broad-spectrum bactericidal activity. They are produced by almost all classes of life to defend against pathogenic infections by interaction with (and disruption of) the bacterial cytoplasmic membrane. Evolution of resistance to antimicrobial peptides has been shown to be difficult because of the requirement for significant rearrangement of the bacterial membrane to confer resistance. However, antimicrobial peptides are difficult to administer at the site of infection because of poor stability. They are often cleared or degraded when delivered; therefore regularly administered high doses are required for efficacy. Because of their potent mechanism of action, the utility of some antimicrobial peptides is further limited by host-toxicity concerns. These challenges could be mitigated by developing a pathogen-specific targeting system. Targeted delivery would also prevent harm to beneficial microbes. We are developing a novel, cell-based drug-delivery platform for antimicrobial peptides in which a therapeutic microbe is engineered specifically to detect a pathogen of interest and deliver antimicrobial peptides in high local concentrations. Delivery of antimicrobial peptides will be achieved by first encapsulating them to produce large amounts of the protein drug and then, upon sensing a specific signal from the pathogen, releasing the antimicrobial peptides by proteolysis and concurrent lysis of the therapeutic microbe. This pathogen-specific targeting system would resolve many of the practical challenges associated with administration of the peptides, including increasing the stability of the payload and reducing off-target effects on other commensal microbes and host cells. This system would be an effective strategic countermeasure against antibiotic-resistant pathogenic bacteria.
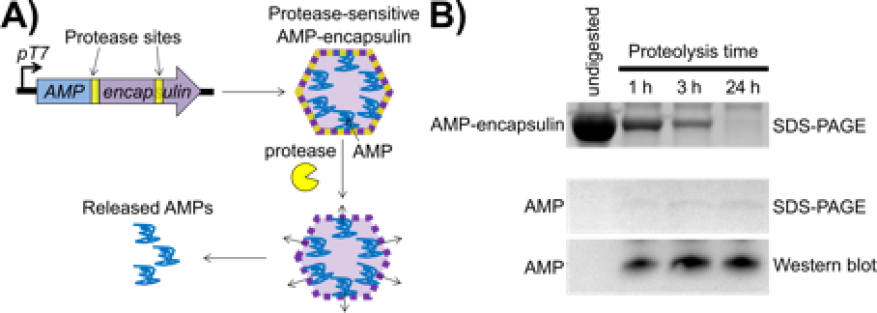
We expect to demonstrate the potential of using a therapeutic microbe to sense a specific pathogen at an infection site, as well as deliver high local concentrations of antimicrobial peptides, thus resolving several barriers to their delivery, including increased stability of the peptides and reduction of off-target effects on host cells and other commensal microbes. We will demonstrate that encapsulation of the toxic antimicrobial peptides will achieve this high expression of peptides in the host therapeutic microbe (see figure). As a proof-of-concept, the probiotic gut organism Escherichia coli will be engineered to serve as the therapeutic microbe to deliver antimicrobial peptides to the opportunistic Pseudomonas aeruginosa bacteria (a multi-drug-resistant pathogen) in the gut environment. The bacteria P. aeruginosa infects human respiratory and gastrointestinal tracts, and is one of the leading causes of hospital-acquired pneumonia. Our specific objectives include production of encapsulated protease-sensitive antimicrobial peptides. Experiments and computational modeling will guide optimization of linking for efficient loading into the compartments and release from the encapsulated structure within the E. coli host organism (and subsequently from the cell itself) by co-expression of the encapsulated system and the appropriate protease. In addition, we will develop release systems in which the encapsulated antimicrobial peptide and protease and lysis systems will be coupled to a specific pathogen-sensing system for P. aeruginosa, which has been thoroughly studied. Finally, we will conduct testing of therapeutic delivery-system efficacy with fluorescent protein reporters using computational modeling of the rates of production and degradation of the system components to guide and improve design, according to current synthetic biology practices. We will then conduct experiments to measure the amounts of antimicrobial peptides released into the medium over time.
Mission Relevance
The development of a therapeutic microbe drug-delivery system is a novel strategic countermeasure against emerging antibiotic-resistant pathogen threats (a national-security concern) that supports the Laboratory's core competencies in bioscience and bioengineering. This work builds upon the Laboratory's portfolio of countermeasures against threats to biosecurity and antibiotic-resistant infections in both military and civilian healthcare environments. It also supports the Department of Energy's goal to transform our understanding of nature and strengthen the connection between advances in fundamental science and technology innovation.
FY17 Accomplishments and Results
In FY17 we (1) worked to develop a system to express three different antimicrobial peptides (LL-37, Apidaecin Ia, and HBCM-2) inside encapsulin compartments by direct covalent attachment; (2) cloned several constructs to express the antimicrobial peptides on the N- or C-terminal ends (amino acid chain terminated by a free amine group or a carboxyl group, respectively) of the encapsulin protein and engineered appropriate protease cleavage sites to potentially break apart the encapsulin shells and release the antimicrobial peptides; (3) began testing the ability of E. coli to express these constructs; (4) identified conditions to release antimicrobial peptides from the encapsulin shells using computational predictions to guide experimental design; and (5) began work on developing a P. aeruginosa-sensing system in E. coli using the LasR quorum-sensing system.