Dan Park (16-LW-055)
Abstract
The ability to detect uranium through environmental sampling is essential for minimizing human exposure, motivating environmental cleanup efforts, and detecting illicit nuclear enrichment programs. However, established technologies for uranium detection are not well-suited for in situ detection of environmental uranium. In our project, we sought to develop an alternative uranium detection strategy using bacteria cells. We took a transcriptomic, molecular genetics, and biochemistry approach—transcriptomics being the study of the complete set of ribonucleic acid (RNA) transcripts produced by the genome, under specific circumstances or in a specific cell, using high-throughput methods, such as microarray analysis. We identified and characterized two novel uranium-responsive gene regulatory systems in the bacterium Caulobacter crescentus, providing the framework for uranium detection using bacterial cells. Leveraging the distinct selectivity profiles of each uranium-responsive regulatory system, we developed a synthetic genetic circuit that integrates signaling input from both regulatory systems and outputs a green fluorescent protein (GFP) reporter in response to concentrations of the uranyl oxycation as low as 700 ppb. This uranium sensing genetic circuitry has enabled a highly selective readout of uranium under laboratory conditions; no cross-reactivity is observed with common environmental metals (e.g., iron, copper, calcium, magnesium, lead, and cadmium) or with the uranium decay chain product thorium. We have also demonstrated that inexpensive, commercially available optic transducer components are sufficient to convert biosensor fluorescence into an electrical current, an important step in the transition of this promising technology from a laboratory-scale towards an automated field portable uranium-detection device.
Background and Research Objectives
Uranium (U) is a highly abundant naturally occurring radionuclide that is critical for nuclear energy and weapon production. Widespread mining and processing of U over the last several decades has resulted in significant U dissemination in the environment (Markich 2002). U is toxic as both a radioisotope and a heavy metal with exposure associated with nephrotoxicity, genotoxicity, reproductive defects, and diminished bone growth (Brugge and Buchner 2011). Accordingly, the Environmental Protection Agency (EPA) has established a maximum contaminant level (MCL) of 30 ppb for U in drinking water. However, ground water concentrations in the U.S. frequently exceed this limit, even in regions lacking anthropogenic activity (Nolan and Weber 2015). To minimize human exposure, motivate environmental cleanup efforts, and detect illicit enrichment programs, there is an imperative need to monitor environmental U concentrations in a robust, cost-effective, and in situ manner.
Established technologies for U detection are not well suited for in situ detection of environmental U. Radiation-based detection methodologies (e.g., gamma and x-ray spectroscopy) are not quantitative in the field and cannot readily distinguish the elemental source of the radiation. Furthermore, these methodologies detect elements of the U decay chain and assume equilibrium with U, an assumption that is unlikely to hold for environmental samples due to mobility of decay daughters. Current analytical methods for direct quantification of U (e.g., inductively coupled plasma mass spectrometry, inductively coupled plasma atomic emission spectroscopy, and kinetic phosphorescence analyzer), while extremely sensitive and selective, are equipment-intensive, costly, and cannot be easily performed in the field. Recently, antibodies and deoxyribonucleic acid (DNA) enzymes with high specificity and selectivity for U have been developed (Liu 2007, Blake 2004), but both systems require sample preparation and costly synthesis or purification steps. In addition, these technologies cannot measure the amount of U that is bioavailable (i.e., present in a form that can exert biological toxicity). Thus, there is a need for the development of alternative sensing technologies that provide sensitive, selective, and cost effective detection of bioavailable U.
Whole-cell biosensors are an alternative and promising choice for field deployment due to their low equipment/energy requirements, low cost, ease of operation, little/no sample processing, and ability to detect bioavailable metal concentrations (Park et al. 2013, Yagi 2007). In response to a stimulus of interest, biosensors output a signal (e.g., fluorescence) that can be readily detected and quantified. This is commonly accomplished through use of an analyte sensing transcription factor that couples signal perception with a gene regulatory response. Through genetic manipulation of these regulatory systems such that the transcriptional regulator activates a reporter protein (e.g., GFP) upon metal perception, whole-cell biosensors have been constructed for a number of environmental toxins, including arsenic, cadmium, lead, mercury, chromium, and copper (Park et al. 2013, Yagi 2007, Dai and Choi 2013, Bereza-Malcolm et al. 2014). However, the construction of a whole-cell sensor of U has been hampered by the absence of characterized U-specific sensing systems.
The objective of our work was to develop a sensitive and selective whole-cell sensor of U and to couple this sensor with optical transducer technology to provide a proof-of-concept demonstration of an integrated U-detection system. Building upon prior studies performed at Stanford (Hu et al. 2005, Hillson et al. 2007) and Lawrence Livermore National Laboratory (Yung et al. 2015), we identified and characterized two novel U-responsive regulatory systems in the bacterium Caulobacter crescentus. Leveraging the distinct selectivity profiles of each U-responsive regulatory system, we developed a synthetic genetic circuit that enabled sensitive and selective detection of the uranyl oxycation. We further demonstrated that this whole-cell sensor could be coupled with inexpensive, commercially available optic transducer components to convert the biosensor fluorescence into an electrical current.
Scientific Approach and Accomplishments
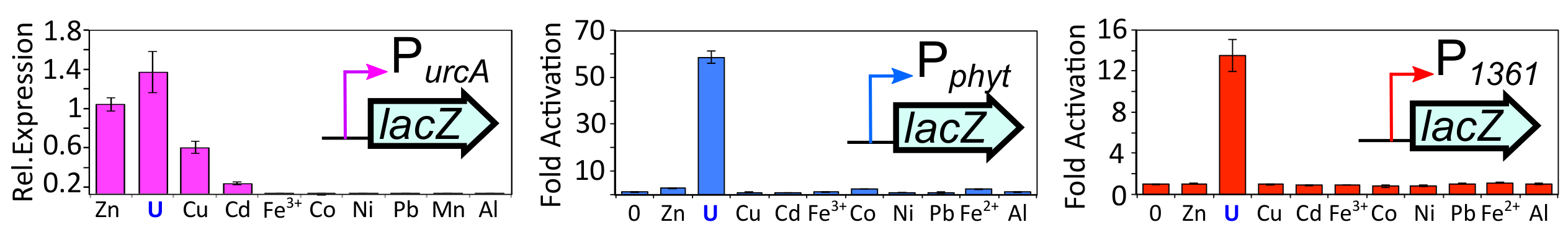
The bacterium Caulobacter crescentus was chosen as a host organism for whole-cell U sensor construction. Caulobacter species tolerate high concentrations of U (Hu et al. 2005, Park and Jiao 2014, Bollmann et al. 2010) and exhibit a robust and specific gene expression response following U exposure (Hu et al. 2005, Yung et al. 2014, Hillson et al. 2007). The highest U-induced gene urcA (uranium response in Caulobacter) has been exploited as a U sensor that can detect micromolar U concentrations in contaminated ground water (Hillson et al. 2007). However, our characterization of the metal specificity of PurcA (promoter of urcA) revealed that although most metals failed to induce PurcA, significant induction was observed with the metal ions zinc, copper, and cadmium in addition to U (see Figure 1; Park et al. 2016). Thus, while PurcA lacks sufficient selectivity to function as a standalone sensor of environmental U, it provides a potential platform for the development of a more selective U sensing circuit.
Through use of a transposon screen, the previously uncharacterized two-component system UzcRS—comprised of the histidine kinase UzcS and the response regulator UzcR—was identified as the metal-dependent activator of PurcA. Using chromatin immunoprecipitation followed by sequencing (ChIP-seq), we identified 40 other UzcRS-regulated promoters in the Caulobacter genome and defined the sequence requirements for UzcRS DNA binding (Park et al. 2016), providing a list of modular genetic parts that were used for subsequent sensor construction.
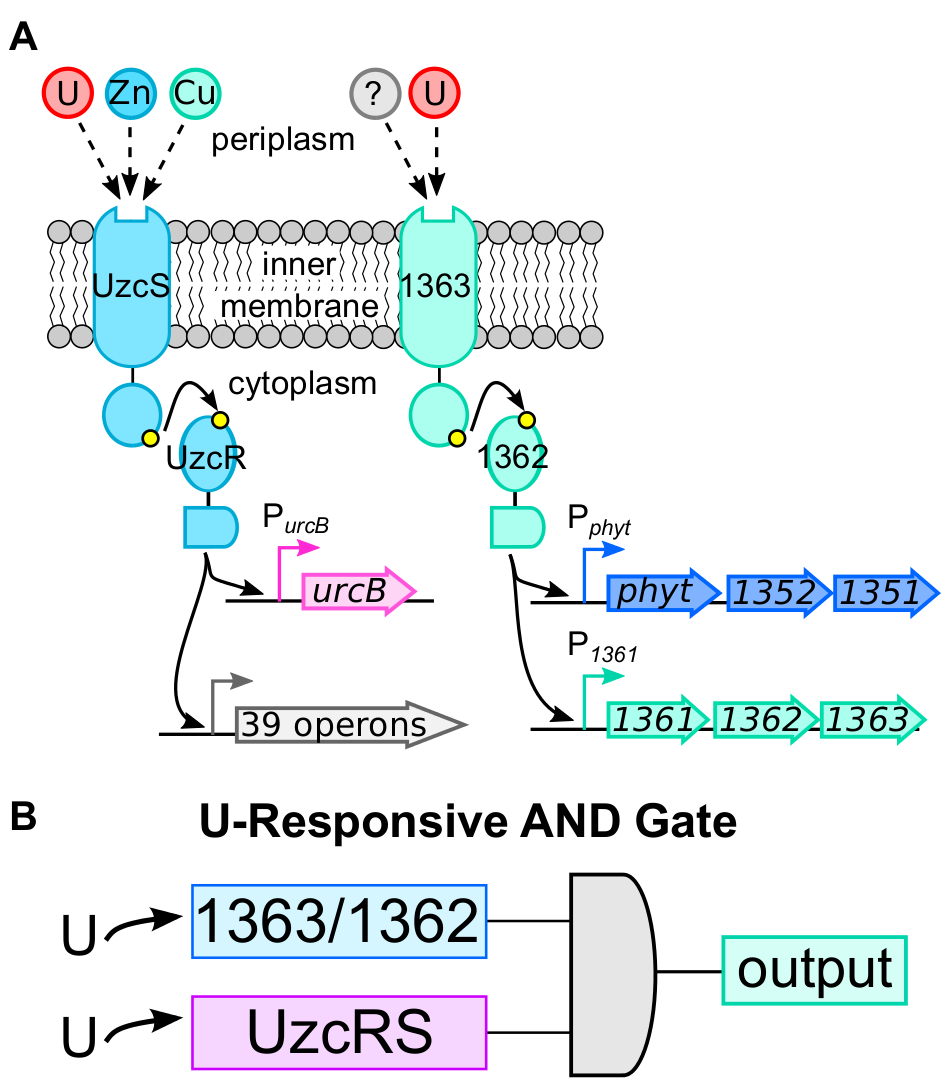
To generate a more selective whole-cell U sensor, we sought to identify a second U-responsive system that functions independently of UzcRS. Using a transcriptomics and molecular biology approach, we identified two additional promoters, Pphyt and P1361, which were responsive to bioavailable U (detection limit ~500 nM) but not to several metals commonly encountered in the environment (see Figure 1). U-dependent induction of both promoters was not dependent on UzcRS (data not shown), indicating that the U-sensing mechanism responsible for induction of Pphyt/P1361 is distinct from that governing UzcRS activity. Through use of a transposon screen, we identified a previously uncharacterized two-component system, comprised of the histidine kinase 1363 and the response regulator 1362, which governs U-dependent activation of Pphyt and P1361. Collectively, these data suggest that C. crescentus possesses two independent, U-responsive two-component systems (Figure 2A).
Given the different selectivity profiles of UzcRS and 1363/1362, we hypothesized that optimal sensor specificity will be accomplished through a dual deployment strategy that incorporates two points of uranyl sensing: (1) 1362/1363 and (2) UzcRS. In this sensor, an output signal will only be produced when both systems are activated, that is, functioning as an AND gate (Bereza-Malcolm et al 2015) (see Figure 2B). Using this AND gate approach, we developed two functional whole-cell U sensors that significantly outperformed the prior art (Hillson et al. 2007) in terms of selectivity for U.
Figure 2. A: Schematic of two independent two-component systems (TCS) that are responsive to uranium (U) in the bacterium Caulobacter crescentus. The UzcRS TCS was characterized as a transcriptional activator of at least 40 operons in response to uranium, zinc, or copper. The 1363/1362 TCS was characterized as an activator of Pphyt and P1361 in response to uranium. In both cases, upon uranium sensing, the histidine kinase (UzcS and 1363) autophosphorylates (adds a phosphate to the kinase) then transphosphorylates (transfers an organic phosphate group) the cognate response regulator (UzcR and 1362, respectively), activating it for DNA binding. The question mark reflects additional, unknown stimuli of 1363. B: Schematic of uranium-responsive circuit design strategy—an AND gate that incorporates signaling input from both UzcRS and 1363/1362.
Sensor I - In series AND gate sensor
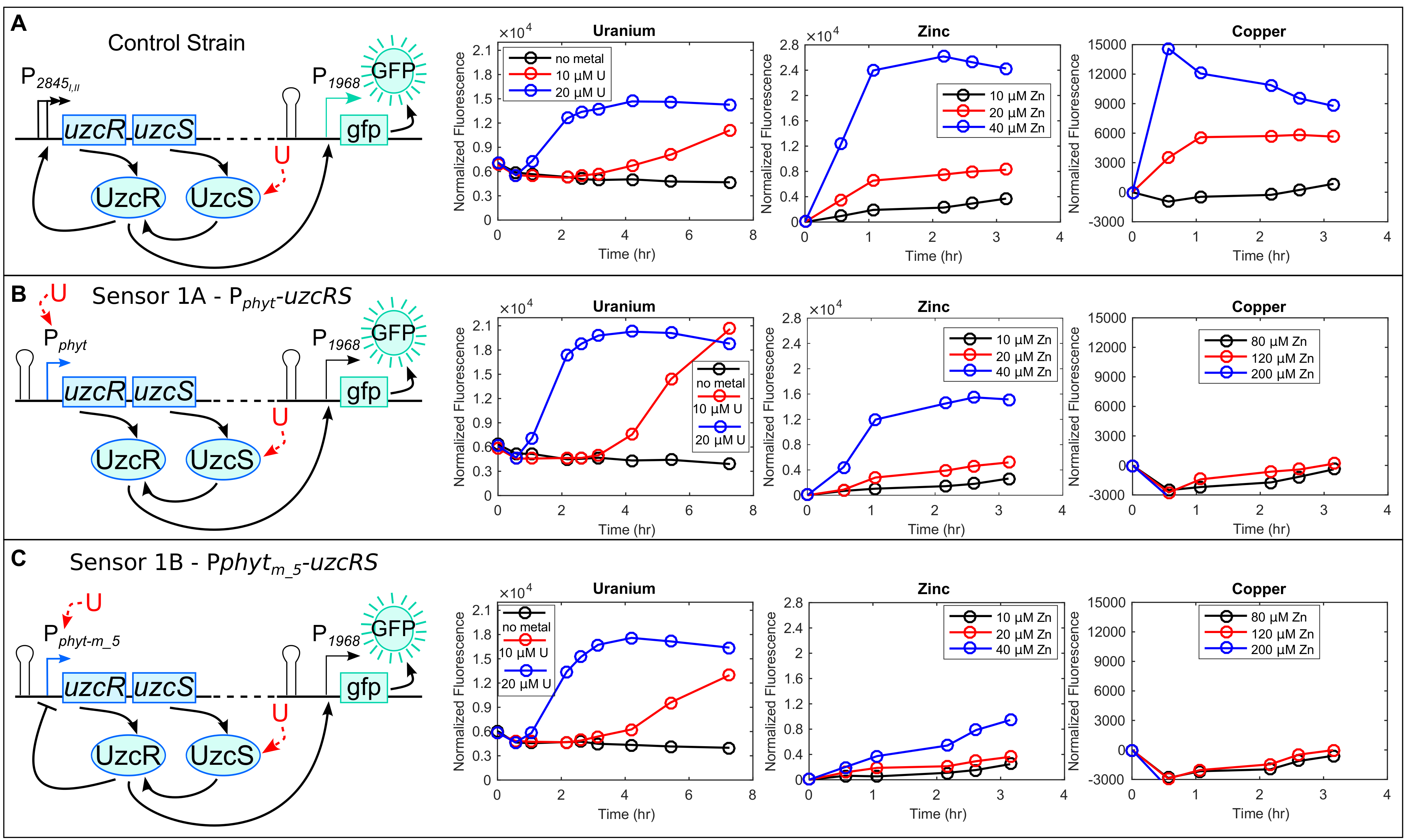
The expression of uzcRS is controlled by two promoters (P1 and P2) that enable sufficient UzcRS protein levels to mediate gene activation in response to the metals uranium, zinc, or copper (Figure 3A). Failing to express uzcRS abolishes PurcA (or any of the other 40 promoters) expression (Park et al. 2016). The overarching goal for Sensor 1 was to make uzcRS expression conditionally dependent on U. To accomplish this, P1 and P2 were replaced with Pphyt such that uzcRS expression requires stimulation by 1362/1363. To monitor the output of this construct (hereafter named Sensor 1A) a UzcR-regulated promoter (P1968) was used to control expression of a fluorescent reporter (gfp; see Figure 3B). As constructed, this circuit requires two points of U sensing for signal output: (1) activation of uzcRS transcription by 1363/1362 and (2) activation of P1968-gfp by UzcR (see Figure 3B). To further improve the specificity of this AND gate, we incorporated a negative feedback loop to further reduce uzcRS expression in the absence of U (Sensor 1B; see Figure 3C). Specifically, a UzcR binding site was placed within Pphyt such that UzcR DNA binding negatively regulates expression of uzcRS. Compared to a control strain lacking modification of the uzcRS promoter, Sensor 1B exhibited a greatly improved selectivity for U (Figure 3A–C). The detection limit of Sensor IB for U is ~700 ppm (data not shown).
Sensor II - In-parallel AND gate sensor
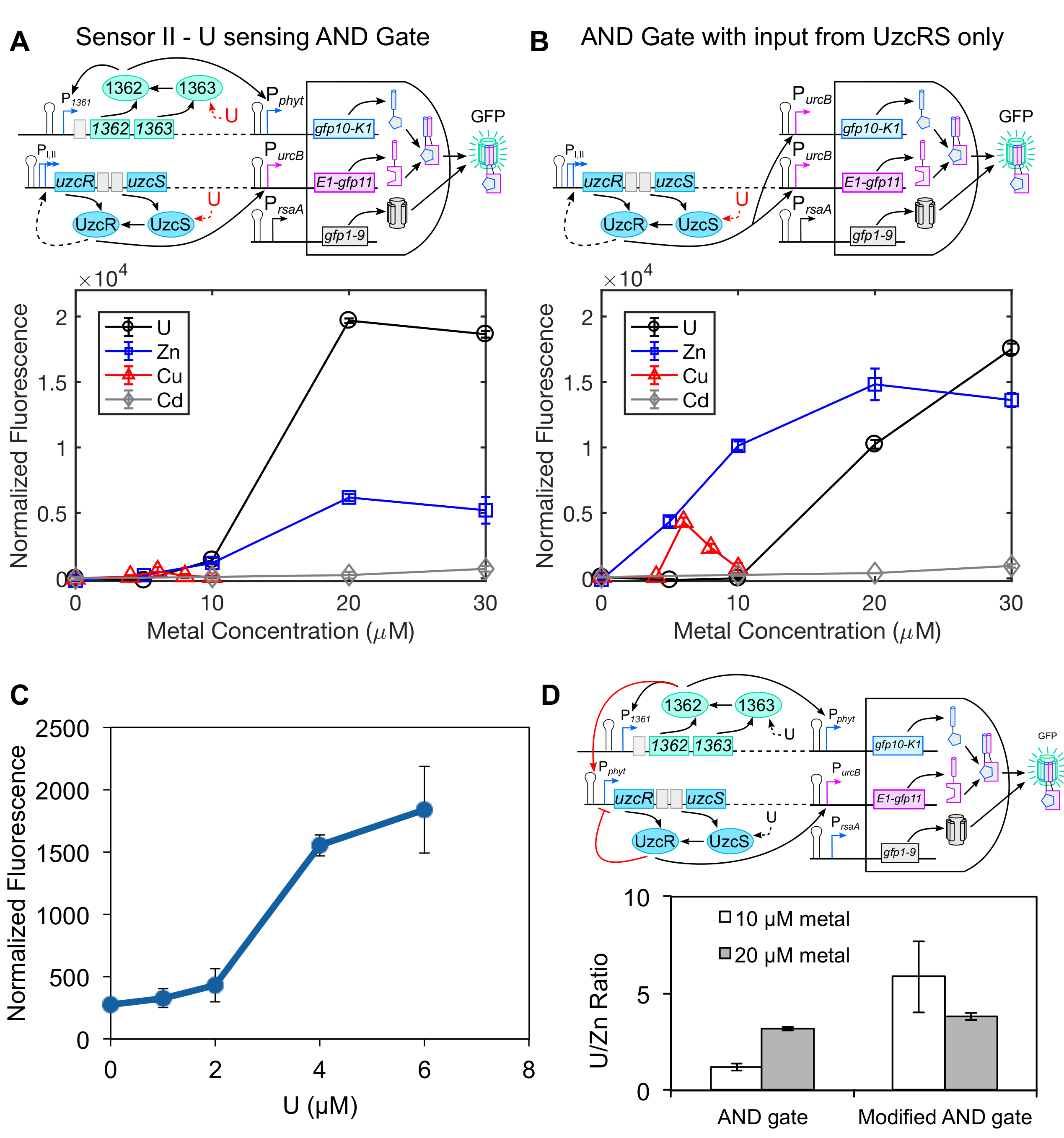
ensor II differs from Sensor I in that it uses 1363/1362 and UzcRS to independently control the expression of two genetic components that are both required for signal output (see Figure 3), providing a potential selectivity advantage over Sensor I. To construct this circuit, we utilized the recently developed tripartite GFP AND gate system (Cabantous et al. 2013) as a template. Pphyt and PurcB (a promoter regulated by UzcRS) were used to drive expression of gfp10 and gfp11, respectively, and gfp1-9 expression was driven by the strong, constitutive rsaA promoter, PrsaA (Fisher et al. 1988), that yields high GFP1-9 levels at all stages of growth. In this circuit, a fluorescent output should only be produced when all three components are expressed and reconstitute GFP. Indeed, cell fluorescence was not observed in cells lacking uzcR or when the 1362 binding site was mutated away from consensus, indicating that U-dependent activation of both the UzcRS and 1362/1363 two-component systems is required for a fluorescence output. Characterization of Sensor II performance revealed a U-concentration dependent increase in fluorescence (see Figure 4A) and a detection limit of ~700 ppb (see Figure 4C). To test the selectivity of Sensor II, a control strain was constructed where expression of both gfp10 and gfp11 are governed by the UzcRS two-component system (see Figure 4B). Compared to the control strain, very minimal fluorescence was observed for Sensor II with zinc and none with copper or cadmium, highlighting the robust specificity for U (see Figures 4A and 4C). We achieved further selectivity against zinc by integrating Sensor I and Sensor II within the same circuit (see Figure 4D).
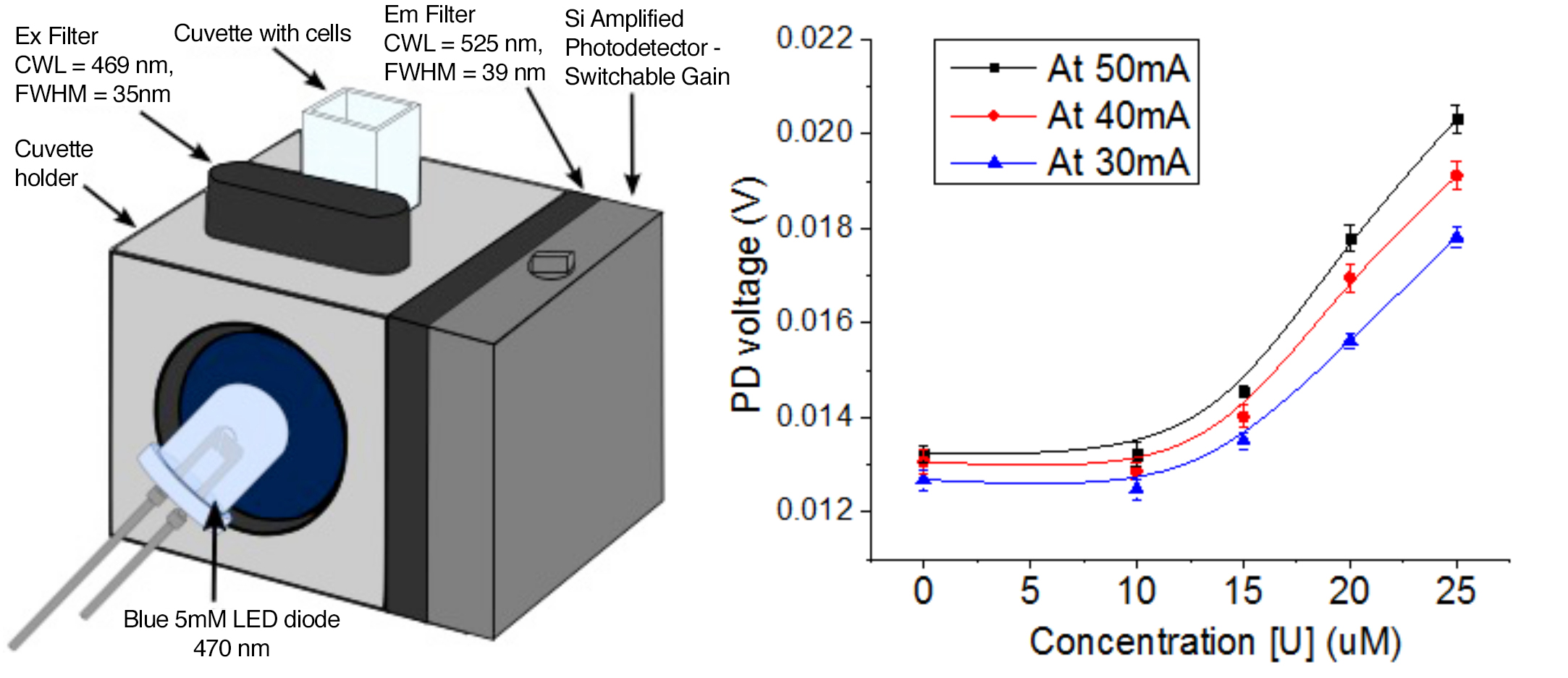
Coupling whole-cell sensor with optical transducer technology
The development of a field-portable biosensor device requires a mechanism to convert the cellular signal into an electric current for readout. As a proof of principle, we built a cuvette-based optical transducer device using commercially available parts from Thorlabs in Newton, New Jersey (see Figure 5). Of particular note was the inexpensive (~50-cent) 5-mm blue light-emitting diode used to excite GFP fluorescence. This setup yielded a U concentration-dependent increase in electrical output, confirming the compatibility of our whole-cell sensors with optical transducer technology (see Figure 5).
Impact on Mission
By providing proof of principle that bacterial regulatory systems can be rewired to enable highly specific and sensitive detection of toxins, our work establishes synthetic biology as a powerful tool for chemical detection at Lawrence Livermore. The expertise gained in this project will benefit future synthetic biology projects aimed at developing cell-based sensors for chemical and biological threat agents. More specifically, the successful development of a whole-cell U sensor provides the foundation for a bacterial-based U-detection platform that will enable a cost-effective, in situ screening method for U. Since U is the most likely signature of an enrichment operation, such a platform will benefit the NNSA's goal of reducing nuclear dangers and the Laboratory's chemical and biological security mission focus area. The flexibility to integrate additional analyte-detection modules (e.g., fluoride) within the genetic circuitry could further target the sensor for nuclear signature detection. Lastly, this work contributes to the Laboratory's basic science mission by providing the first characterization of a U responsive regulatory system in bacteria. These findings increase our knowledge of the fundamental mechanisms that enable microbial survival during environmental stress.
Conclusion
We identified and characterized the metal specificity of two U-responsive regulatory systems (1363/1362 and UzcRS) in Caulobacter crescentus, providing novel insight into how bacteria sense and respond to U in the environment. The integration of both regulatory systems within an AND gate circuit yielded two highly selective whole-cell U sensors. Current research efforts focus on enhancing sensor sensitivity through the incorporation of a genetic signal amplifier module that was identified in FY17. To transition this technology from the proof-of-concept stage (TRL-3) towards a field-portable biosensor device, we will seek additional funding from the Defense Intelligence Agency and NNSA. While we have successfully demonstrated sensor compatibility with optical transducer technology, a critical future task involves the encapsulation and maintenance of the bacterial sensors in an active state within a detection device. As such, we envision fruitful collaborations with Lawrence Livermore researchers who have expertise in microfluidics and device engineering. Ultimately, we envision a use for this technology in anti-proliferation efforts, environmental monitoring and cleanup efforts, and routine monitoring of water sources.
References
Bereza-Malcolm, L. T., et al. 2015. "Environmental Sensing of Heavy Metals Through Whole Cell Microbial Biosensors: A Synthetic Biology Approach." ACS Synth. Biol. 4 (5):535–546. doi: 10.1021/sb500286r.
Blake, II, R. C., et al. 2004. "Novel Monoclonal Antibodies with Specificity for Chelated Uranium(VI): Isolation and Binding Properties." Bioconjug. Chem. 15 (5):1125–1136. doi: 10.1021/bc049889p.
Bollmann, A., et al. 2010. "Isolation and Physiology of Bacteria from Contaminated Subsurface Sediments." Appl. Environ. Microbiol. 76 (22):7413–7419. doi: 10.1128/Aem.00376-10.
Brugge, D., and V. Buchner. 2011. "Health Effects of Uranium: New Research Findings." Rev. Environ. Health 26 (4):231–249.
Cabantous, S., et al. 2013. "A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association." Sci. Rep. 3:2854. doi: 10.1038/srep02854.
Dai, C., and S. Choi. 2013. "Technology and Applications of Microbial Biosensor." Open Journal of Applied Biosensor 2 (3):83–>93. doi: 10.4236/ojab.2013.23011.
Fisher, J. A., et al. 1988. "Transcriptional Analysis of the Major Surface Array Gene of Caulobacter crescentus." J. Bacteriol. 170 (10):4706–4713.
Hillson, N. J., et al. 2007. "Caulobacter crescentus as a Whole-Cell Uranium Biosensor." Appl. Environ. Microbiol. 73 (23):7615–7621. doi: 10.1128/AEM.01566-07.
Hu, P., et al. 2005. "Whole-Genome Transcriptional Analysis of Heavy Metal Stresses in Caulobacter crescentus." J. Bacteriol. 187 (24):8437–8449. doi: 10.1128/JB.187.24.8437-8449.2005.
Liu, J., et al. 2007. "A Catalytic Beacon Sensor for Uranium with Parts-Per-Trillion Sensitivity and Millionfold Selectivity." Proc. Natl. Acad. Sci. USA 104 (7):2056–2061. doi: 10.1073/pnas.0607875104.
Markich, S. J. 2002. "Uranium Speciation and Bioavailability in Aquatic Systems: An Overview." Sci. World J. 2:707–729. doi: 10.1100/tsw.2002.130.
Nolan, J., and K. A. Weber. 2015. "Natural Uranium Contamination in Major U.S. Aquifers Linked to Nitrate." Environ. Sci. Technol. Lett. 2 (8):215–220. doi: 10.1021/acs.estlett.5b00174.
Park, D. M., and Y. Jiao. 2014. "Modulation of Medium pH by Caulobacter crescentus Facilitates Recovery from Uranium-Induced Growth Arrest." Appl. Environ. Microbiol. 80 (18):5680–5688. doi: 10.1128/AEM.01294-14.
Park, D. M., et al. 2017. "Identification of a U/Zn/Cu Responsive Global Regulatory Two-Component System in Caulobacter crescentus." Mol. Microbiol.104 (1):46–>64. doi: 10.1111/mmi.13615. LLNL-JRNL-703325.
Park, M., et al. 2013. "Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery." Sensors (Basel) 13 (5):5777–5795. doi: 10.3390/s130505777.
Yagi, K. 2007. "Applications of Whole-Cell Bacterial Sensors in Biotechnology and Environmental Science." Appl. Microbiol. Biotechnol. 73 (6):1251>–1258. doi: 10.1007/s00253-006-0718-6.
Yung, M. C., et al. 2014. "Shotgun Proteomic Analysis Unveils Survival and Detoxification Strategies by Caulobacter crescentus During Exposure to Uranium, Chromium, and Cadmium." J. Proteome Res. 13 (4):1833–1847. doi: 10.1021/pr400880s.
——— 2015. "Transposon Mutagenesis Paired with Deep Sequencing of Caulobacter crescentus under Uranium Stress Reveals Genes Essential for Detoxification and Stress Tolerance." J. Bacteriol. 197 (19):3160–3172. doi: 10.1128/JB.00382-15.
Publications and Presentations
Park, D. M. 2017. Biosensors for Detecting and/or Neutralizing Bioavailable Uranium and Related U-Sensitive Genetic Molecular Components, Genetic Circuits, Compositions, Methods and Systems. U.S. Provisional Application No.: 62/587,753.
Park, D. M., et al. 2017. "Identification of a U/Zn/Cu Responsive Global Regulatory Two-Component System in Caulobacter crescentus." Mol. Microbiol.104 (1):46–64. doi: 10.1111/mmi.13615. LLNL-JRNL-703325.