Daniel Faissol (17-ERD-036)
Executive Summary
Using a novel combination of high-performance computing, mechanistic-, and data-driven approaches, we are developing predictive biological models, enabling automated hypothesis generation and experiment prioritization for faster development of countermeasures to human health threats and improved predictive capabilities across various national security mission applications.
Project Description
In order to build truly predictive models of complex biological systems, approaches that incorporate both model-driven and data-driven techniques are needed. Mechanistic model-driven approaches, such as molecular dynamics simulations, can only operate at very small time and length scales, and therefore cannot model many systems critical for understanding human diseases. Data-driven approaches, on the other hand, are based on correlations in observed data and are therefore severely limited in their ability to elucidate underlying mechanisms. We are developing a novel methodology for learning interactions within complex biological systems. Our goal is to enable researchers to more efficiently discover biological mechanisms by increasing the automation in hypothesis generation using agent-based models of the biological system. An agent-based model is one of a class of computational models for simulating the actions and interactions of autonomous entities with a view to assessing their effects on the system as a whole. Our approach exploits high-performance computing as well as mechanistic and data-driven modeling approaches to set a path forward for building truly predictive models that allow us to better develop countermeasures against various threats to human health. To accomplish this, we integrate optimization, machine learning, and meta-modeling (the analysis, construction, and development of the frames, rules, constraints, models, and theories applicable and useful to modeling a predefined class of problems) to learn the rules of the agent-based models, which correspond to interactions within the underlying biological system.
If successful, our novel approach will result in an efficient tool for reducing the large space of plausible hypotheses of biological mechanisms. The approach integrates expert knowledge of the biological system together with novel parallel computational methods. Therefore, our study will enable more automation in complex quasi-mechanistic model development, enable mechanistic hypothesis generation to guide which biological experiments are most fruitful to conduct, and lead to a computational framework that bridges the gap between mechanistic and data-driven models. Our primary research efforts focus on five components that are critical for scaling this novel approach to real-world problems: (1) an approximate Bayesian analysis system of statistical inferences based on interpretation of probability, (2) gradient-based optimization, (3) gradient-free optimization, (4) meta-modeling, and (5) temporal feature-based data representations. We aim to leverage existing techniques and develop new ones within each of these components. A major strength of our overall methodology is that the combination of, and interaction among, these disparate approaches helps overcome the weaknesses and limitations of the individual components. Equally importantly, our approach will exploit highly parallelized runs of the agent-based models on high-performance computing platforms.
Mission Relevance
This research directly supports Lawrence Livermore’s chemical and biosecurity strategic focus area in that it is aimed at enabling the development of truly predictive models in biology that will allow us to better develop countermeasures against various threats to human health. Moreover, this research advances the NNSA goal of strengthening the science, technology, and engineering that deals with the nation’s evolving security needs. Given the explosion in agent-based modeling efforts in applications such as cyber networks, vehicle traffic, crowd behavior, economic systems, and financial markets, this research has the potential to improve the predictive capability of models throughout the broader national security mission. Additionally, the expertise gained by developing novel, highly parallelized optimization and statistical techniques directly supports the Laboratory’s core competency in high-performance computing, simulation, and data science.
FY17 Accomplishments and Results
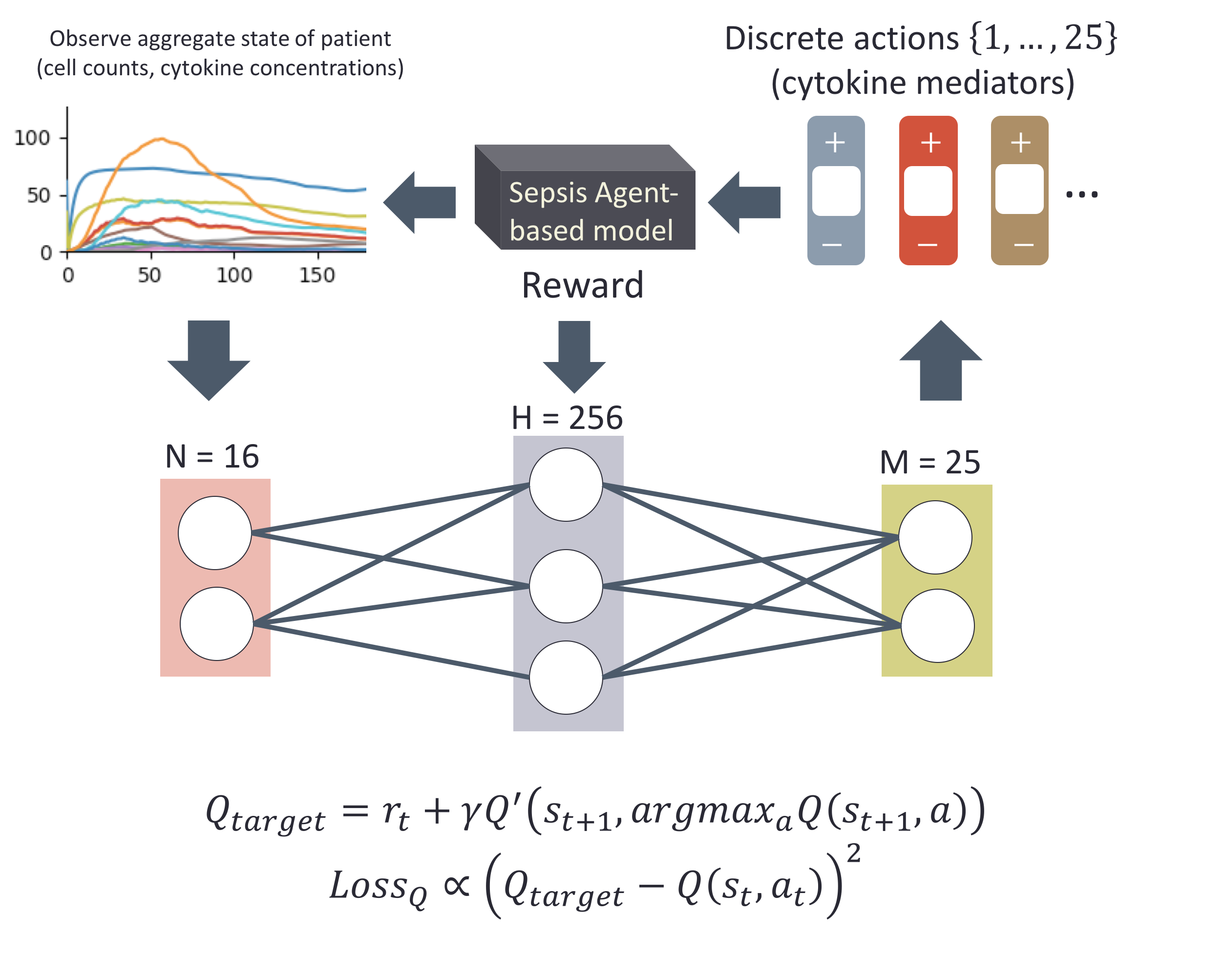
In FY17, we (1) acquired an agent-based model code of the immune response and enhanced it to automatically generate model instantiations with different agent rules; (2) identified the cytokine interaction network as our initial target for learning and identified the relevant data sets; (3) developed and implemented a preliminary gradient-based optimization approach and approximate Bayesian computation rejection algorithm for model parameter calibration using multi-variate time-series data, along with a sparsity-inducing proposal mechanism; and (4) identified an opportunity to exploit the same agent-based model and a reinforcement learning approach to compute a dynamic medical intervention policy capable of “controlling the immune system” via cytokine mediation.