Mimi Yung (15-LW-013)
Project Description
Over the past several years, synthetic biology has utilized the power of genetics to engineer microbes to produce novel products or functions. This work has been enormously useful in producing biofuels and biopharmaceuticals to address the world's energy, medical, and bio-defense needs. In addition, synthetic biology has potential applications in bioremediation via engineered microbes that can degrade toxic chemicals. The core of synthetic biology is the ability to engineer microbes to express novel biosynthetic enzymes or pathways in host cells, such as the Escherichia coli bacteria, that are able to rapidly reproduce and synthesize products of interest. We plan to develop bacterial cell-like "micro-compartments" as novel platforms for synthetic biology. These micro-compartments are bacterial organelles that are made of a protein shell that surrounds and encloses various enzymes. We will bioengineer the micro-compartments to encapsulate heterologously expressed proteins in host cells to prevent cytotoxicity of the protein or reaction intermediates and improve reaction efficiency, which are two major bottlenecks preventing the expression of novel proteins and functions in heterologous hosts. Heterologous expression refers to the synthesis of a gene or part of a gene in a host organism that does not naturally have this gene. As proof of concept, we will explore two applications: the expression of the cytotoxic antimicrobial protein CistronA in bacterial micro-compartments for biopharmaceutical and biodefense production, and the expression of the hydrocarbon oxidizer ToMO (toluene/o-xylene monooxygenase) in bacterial microcompartments for the bioremediation of trichloroethylene, commonly used as an industrial solvent.
We expect, if successful, to demonstrate that engineered bacterial micro-compartments are able to alleviate host cell cytotoxicity of proteins or reaction intermediates, thereby enhancing non-native protein expression and improving reaction efficiency, which would be major advancements in synthetic biology. We will develop a novel platform for synthetic biology based on the encapsulation of heterologously expressed proteins in bacterial micro-compartments through the use of computational modeling; DNA cloning; protein expression, purification, and characterization; and transmission electron and fluorescence microscopy. We expect to demonstrate that this system has advantages compared to conventional protein expression systems because it can be used to express proteins that are normally cytotoxic to host cells, as well as improve reaction efficiency for enzymes that produce cytotoxic intermediates. This will have broad utility for the production of antimicrobial proteins, such as endolysin enzymes responsible for cleaving cell walls for viral reproduction, which have important biodefense applications. The development of this platform will aid not only biopharmaceutical production of cytotoxic proteins relevant to biodefense, but also biofuels production and bioremediation involving heterologous protein expression. The use of bacterial micro-compartments could be applied in the future to complex bioremediation pathways, such as the degradation of polychlorinated biphenyl organic industrial pollutants.
Mission Relevance
If successful, bacterial micro-compartments can be leveraged for the heterologous expression of many other proteins or biosynthetic pathways that are critical for antimicrobial drug development, biofuels production, and the cleanup of toxic chemicals in the environment. Additionally, the technology will have broad impacts in many fields of biology, which rely on the production of active protein for a diverse range of studies, including those in medicine and biodefense, and supports the Laboratory's core competency in bioscience and bioengineering.
FY16 Accomplishments and Results
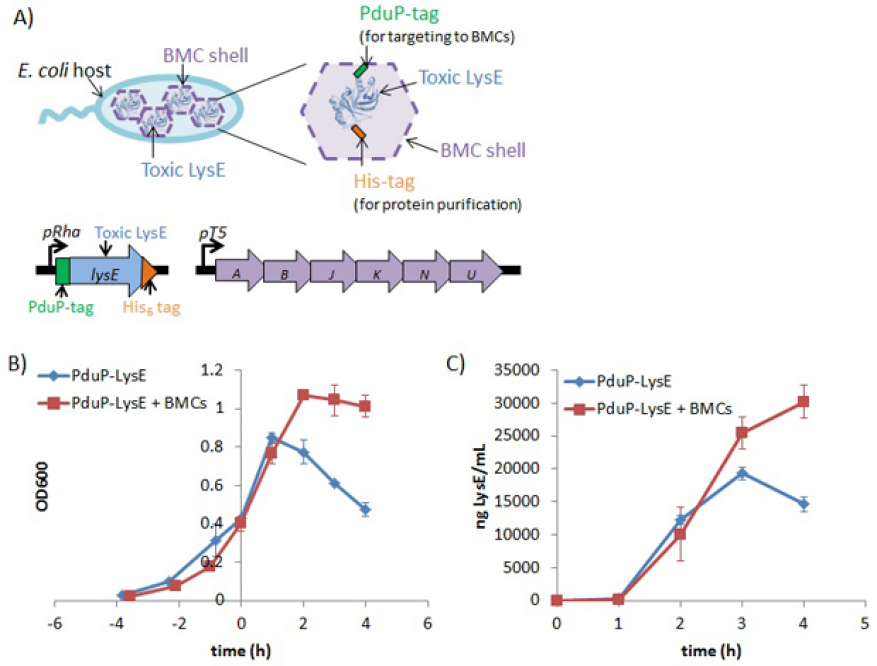
We chose to focus on conducting a basic science analysis of how LysE, a difficult-to-express cytotoxic protein, is packaged inside bacterial micro-compartments, given initial problems finding a suitable toxic protein for encapsulation. Therefore, in FY16 we (1) identified optimal conditions to express the viral protein LysE in bacterial micro-compartments, using an N-terminal (start of a protein or polypeptide terminated by an amino acid with a free amine group) bacterial micro-compartment targeting protein tag (PduP tag); (2) demonstrated co-expression of bacterial micro-compartments with PduP and LysE that resulted in approximately twofold improvements in both host bacterial growth and PduP–LysE production levels, compared to cells expressing only PduP and LysE (see figure); and (3) successfully achieved affinity purification of PduP–LysE from cells producing bacterial microcompartments in the presence of mild detergent.
Publications and Presentations
- Yung, M. C., et al., Engineering bacterial microcompartments (BMCs) to shield toxicity during protein expression and purification. (2015). LLNL-POST-672368.