Nicholas Be (17-ERD-031)
Executive Summary
We are combining advanced capabilities in DNA sequencing, protein detection, and bioinformatics analysis with computational expertise in network modeling to develop a comprehensive portrait of combat wounds and methods for improving care. Our data will be integrated in a first-of-its-kind network model to profile the interaction of microbial and human factors and their impact on healing.
Project Description
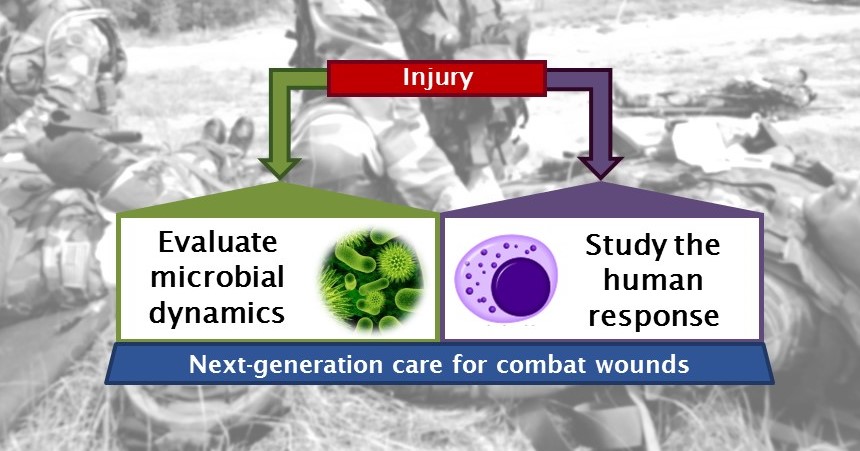
The healing process in traumatic combat wounds, acute civilian wounds, and chronic non-healing wounds is complex. Combat wounds are unique in severity, systemic impact, injury environment, and treatment challenges. The decision to apply specific care should be based on a complete characterization of this process (see figure). However, the functional interaction of human and microbial factors in wound healing is not well understood. A wide range of interacting factors are involved in response to a traumatic wound, including human proteins such as pro-inflammatory cytokines. Infection represents a substantial complication within the wound microenvironment. Colonization of traumatic wounds by microorganisms, referred to as the wound “bioburden,” is associated with an increase in detrimental pro-inflammatory and degradative factors, leading to a systemic response that impedes healing. A more accurate assessment of wound progression is critical to reducing patient morbidity. We are applying deep metagenome DNA sequencing (the study of genetic material recovered directly from environmental samples) for high-resolution assessment of the microbial species, strains, and genes that impact healing in a clinical cohort of combat-injured patients. Our microbial analysis is performed along with a parallel assessment of human immune factors known to be involved in wound healing. All data will be integrated in a computational network model that predicts dependencies between all factors, facilitating more accurate prognoses and treatment guidance for patients. The resultant comprehensive survey will combine a high-resolution microbial species and gene census with a portrait of the human immune response to injury and infection. Simultaneous assessment of host and microbial factors and their interdependencies would facilitate a deeper understanding of wound healing.
We are combining advanced capabilities in DNA sequencing, protein detection, and bioinformatics analysis with computational expertise in network modeling. Through established partnerships with collaborators in combat medicine, we are applying these strengths to a unique clinical panel of wounds from combat-injured service members to create a comprehensive population profile of the wound bioburden. We will provide species and strain-specific identification of all microbial organisms present within infected combat wounds and construct a complete profile of the wound microbial metagenome. We will also assess the gene content of the microbial population to identify the potential of these organisms to negatively impact the healing process and interrogate the human expression profile and protein content in combat wounds. A panel of human proteins and genes known to impact wound healing will be determined in the same samples characterized for microbial content. Finally, we will build a network model of the wound microenvironment. Our data will be integrated in a functional network model to profile the interaction of microbial and human factors and their impact on wound healing, providing clinicians with critical tools to administer personalized intervening care.
Mission Relevance
This project supports the NNSA goal of strengthening its science, technology, and engineering base. Our model has wide-ranging applications in traumatic military and civilian medicine, supporting the Laboratory's core competency in bioscience and bioengineering. Addressing Livermore's priorities to develop and experimentally validate advanced efforts in computation supports the core competency in high-performance computing, simulation, and data science.
FY17 Accomplishments and Results
In the first several months of this project, we have been curating and preparing for analysis a large panel of clinical samples and optimizing molecular methods for detection and characterization of clinically relevant wound pathogens. In FY17 we (1) established the project framework, (2) completed and analyzed a whole-genome sequencing pilot experiment, (3) established the required sequence depth for assessing metagenomic complexity, and (4) established a comparative assessment of techniques for microbial nucleic acid enrichment, providing a baseline of parameters for guiding and improving performance in future experiments.