Monica Borucki (13-ERD-020)
Abstract
Novel pathogens may circulate in a population for years prior to detection, severely hampering prompt and appropriate treatment and containment. Recent advances in characterizing microbes include microarrays, metagenomics, and next-generation sequencing. These techniques rely on detection of the microbe genome, which exists in much smaller amounts than the host genome. This research project aims to develop rapid techniques that separate pathogen nucleic acid from host genetic material to enable detection of novel pathogens from complex samples. Methods will be developed to rapidly isolate pathogen genetic material captured by the host's Toll-like receptors, which are a class of proteins that play a key role in the innate immune system, and immunoglobulin M, which is a basic antibody. The captured genomes will be characterized using metagenomics (the study of genetic material recovered from environmental samples). In addition to pre-symptomatically detecting pathogens before an outbreak, this technique could be used to screen archived blood samples from all over the world to identify the genotypes circulating before the outbreak and pinpoint the genetic changes that led to the outbreak. These data could also be used to generate a database of early-stage infections for determining which viruses routinely infect humans and the geographical location of each virus, as well as providing insight into interactions between a host and pathogen.
We expect to produce assays in which the extraction and purification of viruses or viral ribonucleic acid bound to immunoglobulin M and Toll-like receptors, respectively, from pathogen-infected mice allows microbial nucleic acid to be concentrated away from host nucleic acid. Once the pathogen genome is isolated in this way, it will permit efficient and in-depth metagenomic characterization of the pathogen genome. We will focus on detecting ribonucleic acid viruses, because these are the most challenging group of pathogens to detect. Once tested and optimized on control samples, the methods we develop in this project can then be tested in human biological-surveillance efforts such as testing serum and nasal samples from military personnel returning from overseas deployment for the presence of exotic microbes.
Mission Relevance
In support of the Laboratory's core competency of bioscience and bioengineering, this project will enable the potentially pre-symptomatic detection of emerging and bio-engineered viruses, thereby providing precious time for the rapid development of therapeutics against these threats. This method could also be used to screen archived samples to create a genomic database for identifying the genetic changes that lead to outbreaks and for other insights into host–pathogen interactions.
FY15 Accomplishments and Results
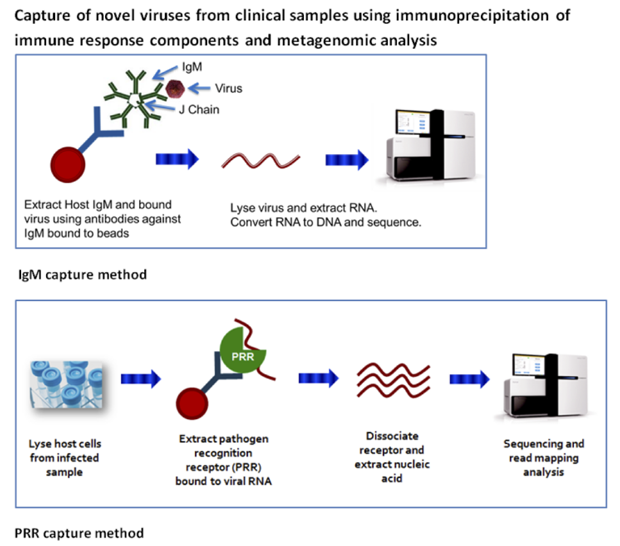
In FY15 we (1) tested immunoprecipitation assays for detection of viral-bound multiple-pathogen recognition receptors TLR-3, MDA5, and RIG-I on clinical samples; (2) tested, with a collaborator at the National Institutes of Health, blood samples from macaque monkeys infected with hantavirus or Nipah virus; (3) tested the yield using metagenomics—in each case, viral ribonucleic acid was detected, but yield was low; (4) added a nonspecific amplification step to increase yield; (5) tested immunoprecipitation assays using the immunoglobulin M antibody and J chain (a protein component of immunoglobulin M), using lung lavage from mice infected with Sendai virus and nasal samples from camels infected with the Middle East Respiratory Syndrome coronavirus; (6) used polymerase chain reaction, a molecular biology technology that amplifies a single copy or a few copies of a piece of deoxyribonucleic acid or ribonucleic acid across several orders of magnitude, to determine that the assays were successful (metagenomics verification is pending); (7) performed deep sequencing on samples of the Middle East Respiratory Syndrome coronavirus to detect the presence of viral genetic variants; and (8) initiated a collaboration with the California National Primate Research Center located at the University of California, Davis, for testing of nonhuman primates for viral pathogens.
Publications and Presentations
- Borucki, M., et al., Middle East respiratory syndrome coronavirus intra-host populations are characterized by numerous high frequency variants. (2015). LLNL-ABS-678358.