Mimi C. Yung (15-LW-013)
Abstract
The objective of this project was to demonstrate that bacterial microcompartments (BMCs) found in nature can be re-directed for novel synthetic-biology applications. BMCs are bacterial “organelles” (i.e., a specialized part of a cell with a specific function) in which enzymes comprising a biosynthetic pathway are encapsulated in a proteinaceous shell in order to, in part, prevent toxicity of reaction intermediates. As a proof-of-concept, we engineered BMC systems to enhance recombinant expression (i.e., production) of a toxic viral protein, lysis protein E (E), from bacteriophage phiX174 in the host bacterium Escherichia coli. We hypothesized that targeting E to the interior of BMCs for encapsulation would shield the toxicity of E and thus improve its expression in Escherichia coli. In this case, E was fused with various N-terminal BMC targeting tags and co-expressed with appropriate BMC shell proteins that associate with the tags and are required to form BMCs. A PduP1-18-tagged E system (PduP being a protein) appeared to achieve the highest expression of E. Co-expression of Pdu BMC shell proteins with PduP1-18-E increased its expression by approximately 50 percent. Affinity purification of PduP1-18-E yielded 270 micrograms (µg) of PduP1-18-E per liter of induced culture. Removal of the PduP1-18-tag via proteolysis resulted in a final yield of 200 µg of E per liter of induced culture, a nearly seven-fold improvement compared to values documented in prior reports. These results demonstrate improved expression of E via re-directed BMC systems and ultimately higher E-purification yields. Our strategy demonstrates the utility of engineered BMC systems for novel synthetic-biology applications.
Background and Research Objectives
Heterologous expression of proteins in host organisms plays a central role in biochemistry and synthetic biology. Protein expression, however, can be challenging, especially if the protein of interest is toxic to the host organism (Rosano and Ceccarelli 2014; Keasling 2012). One potential method to overcome toxicity is to encapsulate the toxic protein within other shell proteins, in order to sequester it from interactions in the host organism. Many bacteria have evolved a mechanism to compartmentalize proteins comprising a biosynthetic pathway inside self-assembling protein shells called bacterial microcompartments (BMCs) (Chowdhury et al. 2014). BMCs are thought to prevent toxicity of reaction intermediates in the biosynthetic pathway, as well as increase the pathway-reaction efficiency by increasing the local concentration of enzymes and substrates within the compartments. Re-directing BMC shell proteins to encapsulate a toxic protein has the potential to shield the toxicity of recombinant proteins, enabling downstream applications such as purification and enhanced activity for a desired function.
As a proof-of-concept, we attempted to encapsulate lysis protein E from bacteriophage phiX174 (hereafter abbreviated as "E") inside recombinant BMCs in Escherichia coli to improve expression and downstream purification of this toxic protein. E is a 91-residue membrane protein that is responsible for bacterial cell destruction (lysis) by phiX174 bacteriophage. It inhibits host cell-wall biosynthesis and thus is extremely toxic to Escherichia coli (Bernhardt et al. 2000). Over-expression of E in Escherichia coli has been difficult, as it induces rapid cell death within 15 minutes, resulting in poor E yields. The best reported purification of E resulted in a yield of 27 µg of E per liter of culture with 84 percent purity (Zheng et al. 2009).
We targeted E to the inside of recombinant BMCs derived from 1,2-propanediol utilization (Pdu) and ethanolamine utilization (Eut) systems found in enteric bacteria (Chowdhury et al. 2014) (Figure 1). Several recent studies have identified components to successfully re-direct non-native proteins to the interior of Pdu and Eut BMCs, including the genes responsible for forming empty BMCs as well as short N-terminal sequences that interact with BMC shell proteins for interior targeting (Fan et al. 2012; Lawrence et al. 2014). E was fused to different N-terminal BMC targeting tags and co-expressed with appropriate BMC shell proteins. Addition of N-terminal targeting tags onto E allows it to be expressed at relatively high levels, while co-expression of BMC shell proteins with E can increase expression by approximately 50 percent with a PduP1-18-tagged/Pdu BMC system. Purification by nickel-affinity chromatography (Ni-NTA) followed by proteolytic cleavage (i.e., the process of breaking the peptide bonds between amino acids in proteins) of the PduP1-18-tag yielded 200 µg of E per liter of induced culture (approximately 80 percent pure), an almost seven-fold improvement in yield compared to those documented in prior reports (Zheng et al. 2009).
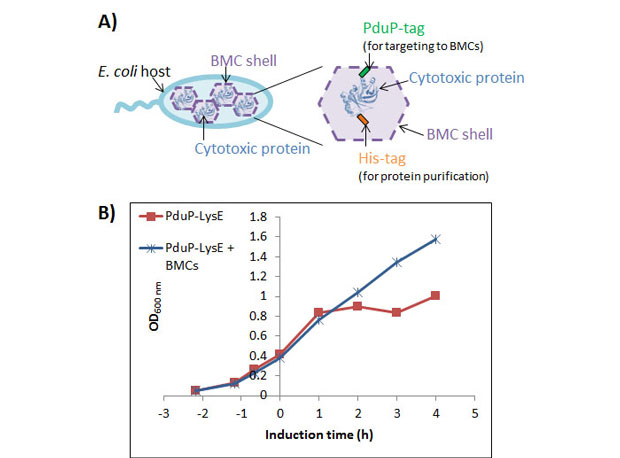
Figure 1. Top: Schematic diagram of strategy for expression of toxic lysis protein E (E or Toxic E) in bacterial microcompartments (BMCs). E is co-expressed with BMC shell proteins in a bacterial host, Escherichia coli. Association of E with BMCs (BMC shell) shields its toxicity to the host, allowing for high levels of E production. E is fused to an N-terminal tag (PduP1-18-tag) for targeting to BMCs as well as a hexa-histidine tag (His6-tag) for later protein purification. Bottom: Genes required for expression of the E construct (blue) and BMC shell proteins (purple).
Scientific Approach and Accomplishments
Different BMC Systems Alter E Protein-Expression Levels
In order to target E to the interior of BMCs, E was fused with known N-terminal BMC targeting tags (18-19 amino acids) and expressed in combination with appropriate BMC shell proteins from Eut and Pdu operons in Salmonella enterica, which were previously shown to produce empty, recombinant BMCs (Choudhary et al. 2012; Sargent et al. 2013; Fan et al. 2010). Escherichia coli strains were constructed to express the following: EutC1-19-E + EutSMNLK; EutC1-19-E + EutS; and PduP1-18-E + PduABJKNU (Figure 2A). The gene for each E construct was expressed from a rhamnose inducible promoter (pRha), which was chosen for tunable expression (expression level varies with rhamnose-inducer concentration). The BMC shell proteins were expressed from a T5 isopropyl-beta-D-thiogalactoside (IPTG) inducible promoter. All E constructs were designed to express a C-terminal His6-tag for later purification of the protein. Factor Xa and thrombin protease-cleavage sites were engineered appropriately for potential cleavage of the BMC targeting and His6 tags, respectively.
To compare expression of the tagged-E constructs, the different Escherichia coli strains were grown in Luria-Bertani (LB) growth medium and induced under one of the following conditions: IPTG only (BMC), rhamnose only (E), or rhamnose and IPTG co-induction (E + BMC). BMC expression was induced at the start of growth, while E construct was induced once cells reached an optical density at 600 nm (OD600) of 0.4 (about three hours after the start of growth). The timing of BMC induction was chosen to be well before E induction to ensure that enough BMC protein was available for encapsulation once E was expressed. Growth rates under the different induction conditions for each E/BMC combination strain were monitored by OD600 (Figure 2B), and after four hours of post-rhamnose induction, the relative expression level of E protein for equivalent volumes of culture was determined by Western blot analysis (Figure 2C).
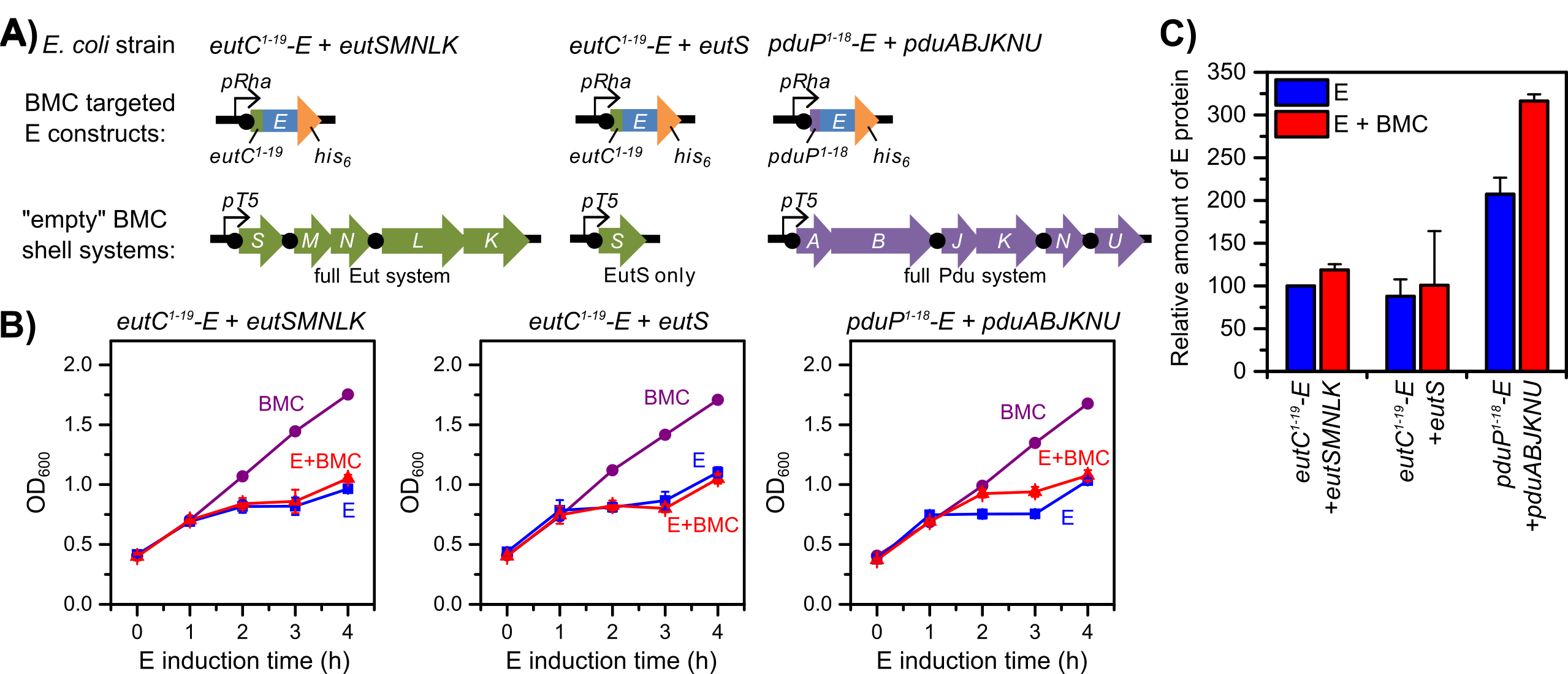
Figure 2. Different re-directed bacterial microcompartment (BMC) systems alter protein expression levels of toxic lysis protein E (E). The PduP1-18-E + PduABJKNU system is the only system to show improved growth and E expression (approximately 50 percent higher) under E + BMC co-induction, compared to E-only induction, demonstrating toxicity shielding of PduP1-18-E by BMC co-induction. A: Genetic scheme for different re-directed BMC systems. B: Growth curves (optical density, OD600 vs. E induction time in hours, h) for each strain under different induction conditions. BMC-only induction (purple circles); E-only induction (blue squares); and E + BMC co-induction (red triangles). C: Relative amounts of E protein based on Western blot intensities (compared to EutC1-19-E + EutSMNLK, E only). E-only induction (blue bars); E + BMC co-induction (red bars). Error bars represent triplicate cultures.
For all strains, the BMC-only induction condition showed normal growth for Escherichia coli, demonstrating that expression of the different BMC shell proteins alone is not toxic to the cells. In contrast, the E-only induction conditions showed significant toxicity (manifested as an arrest or decline in OD600) due to expression of the E construct. Interestingly, the PduP1-18-E/Pdu BMC strain (pPduP1-18-E + PduABJKNU) was the only strain to exhibit improved growth under E + BMC co-induction compared to E-only induction. This result indicated some toxicity shielding due to co-expression of the full Pdu BMC system. The maximum growth difference was observed at three hours, where the E + BMC co-induction culture had approximately 20 percent higher OD600 than the E-only culture. Western blot analysis (used to separate and identify proteins) revealed that the PduP1-18-E protein level in the E + BMC culture was 149 (± 15) percent higher than the respective E-only culture. Given the high PduP1-18-E expression level and toxicity shielding observed with the PduP1-18-E/Pdu BMC system, it was chosen for further study and optimization. By altering the induction timing and concentrations of IPTG and rhamnose inducers, the expression level of PduP1-18-E was optimized to ultimately produce approximately 10 mg/L of culture.
Toxicity Shielding is Associated with Co-Expression of Pdu BMC Shell Proteins
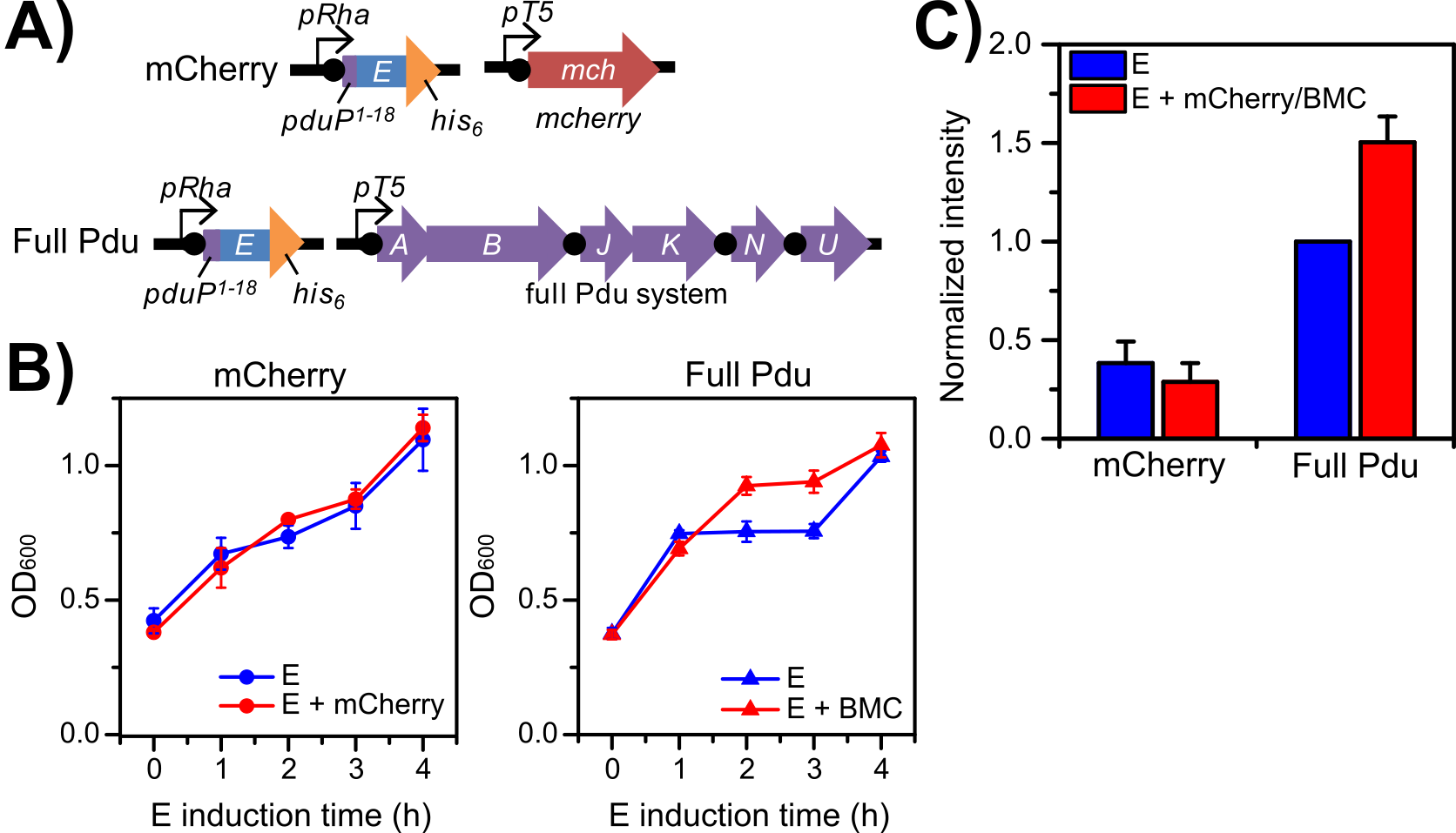
In order to confirm that the effect observed in the PduP1-18-E/Pdu BMC system is associated with co-expression of BMC shell proteins, we prepared a control strain, where PduP1-18-E can be co-expressed with mCherry instead of PduABJKNU. Growth and PduP1-18-E expression in this PduP1-18-E/mCherry control strain was compared to the PduP1-18-E/Pdu BMC strain (Figure 3). Growth curves for the control strain revealed no growth difference between E + mCherry co-induction versus E-only induction and demonstrated a similar degree of toxicity as the growth curve for the PduP1-18-E/Pdu BMC strain under E-only induction (Figure 3B). In contrast, the PduP1-18-E/Pdu BMC strain under E + BMC co-induction showed improved growth at two hours and three hours post-induction. Western blot analysis of equivalent volumes of culture after four hours post-induction (Figure 3C) also showed no significant difference in PduP1-18-E expression in the PduP1-18-E/mCherry control strain under E + mCherry co-induction versus E-only induction. In contrast, PduP1-18-E levels in the PduP1-18-E/Pdu BMC strain under E + BMC co-induction were 50 (± 13) percent higher than under E-only induction. These results indicate that toxicity shielding in growth and higher overall PduP1-18-E yield are associated with co-expression of BMC shell proteins.
Figure 3. PduP1-18-E toxicity shielding is associated with co-expression of Pdu bacterial microcompartment (BMC) proteins. Enhanced growth and PduP1-18-E levels are only observed in a Pdu BMC co-expression system, and not with an mCherry co-expression system. (A) Genetic scheme for the PduP1-18-E/mCherry control strain (mCherry) and the PduP1-18-E/Pdu BMC strain (Pdu BMC). (B) Growth curves for each strain under different induction conditions (optical density, OD600 vs. E induction time in hours, h). E-only induction (blue shapes); E + mCherry or BMC co-induction (red shapes). (C) Relative comparison of amounts of PduP1-18-E protein levels based on semi-quantitive Western blot signal intensities (compared to Pdu BMC, E-only). E-only induction (blue bars); E + mCherry or BMC co-induction (red bars). Error bars represent triplicate cultures.
PduP1-18-E Co-Purifies with Isolated BMCs
We next examined whether PduP1-18-E is associated with recombinant BMCs in vivo. BMCs were isolated from PduP1-18-E/Pdu BMC cells co-induced with 0.5 mM IPTG (for BMC production) and 0.1, 0.2, or 0.5 mM rhamnose (for PduP1-18-E production) for three hours according to previously documented methods (Sinha et al. 2012). As a positive control, BMCs were also isolated from Escherichia coli cells co-expressing a PduP1-18-tagged mCherry (PduP1-18-mCherry) and Pdu BMCs. To determine the amount of contaminating PduP1-18-E (that is non-specifically purified during the BMC isolation and is not BMC-associated), a mock isolation was also performed using Escherichia coli cells only expressing PduP1-18-E.
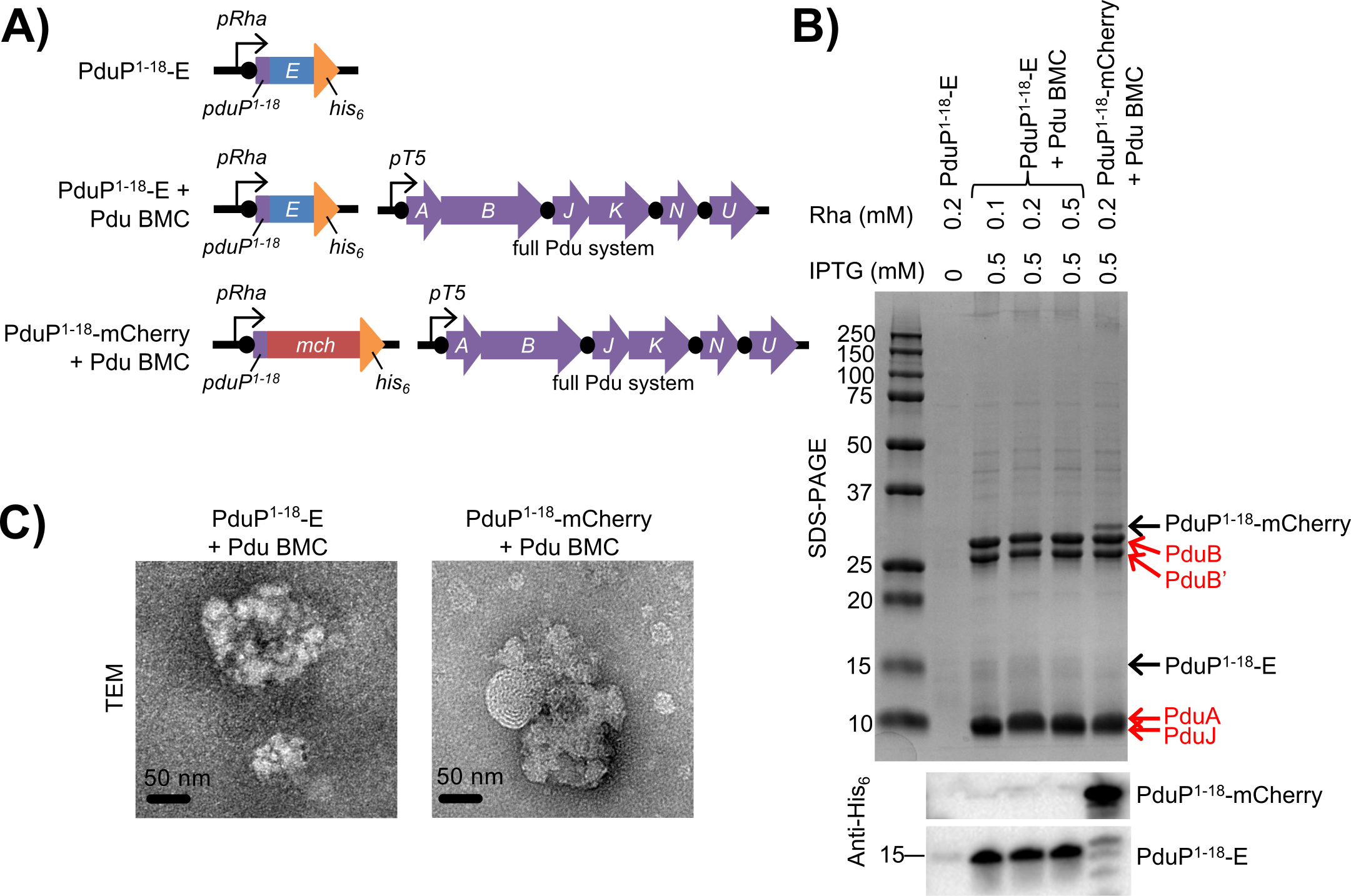
Isolated BMCs were analyzed by SDS-PAGE, anti-His6 Western blot, and transmission electron microscopy (TEM) (Figure 4). As expected, isolated BMCs across samples primarily contain shell proteins corresponding to sizes for PduB, PduB’, PduA, and PduJ by SDS-PAGE (Figure 4B). TEM analysis revealed that isolated BMCs from the different strains are morphologically similar with irregular shapes of up to approximately 200 nm in diameter (Figure 4C), similar to other isolated recombinant BMCs observed in prior studies (Parsons et al. 2008; Sargent et al. 2013). Furthermore, SDS-PAGE and Western blot analysis revealed that PduP1-18-mCherry (~31 kDa) or PduP1-18-E (approximately15 kilodaltons, kDa) was co-isolated with appropriate Pdu BMC samples. Very little contaminating PduP1-18-E was found in the mock BMC isolation from the PduP1-18-E only strain, less than 0.8 percent of that isolated from the PduP1-18-E/Pdu BMC strain (Figure 4B). Interestingly, the amounts of PduP1-18-E co-isolated per mg of total BMC protein were very similar in all three BMC samples from the PduP1-18-E/Pdu BMC strain, despite the differences in rhamnose induction concentrations: 12-14 µg of PduP1-18-E per milligram of total BMC protein. This result suggested that PduP1-18-E was fully loaded into the isolated BMCs and increasing rhamnose concentrations, corresponding to increasing expression of PduP1-18-E, did not improve load.
However, of the total expressed PduP1-18-E in the cell lysate, only approximately one percent was found to be co-isolated with BMC samples from the PduP1-18-E/Pdu BMC strain. In the positive control, the amount of PduP1-18-mCherry co-isolated with BMCs was similarly low at 0.25 percent of the total PduP1-18-mCherry in the cell lysate. Given the size heterogeneity and irregular shapes of the isolated BMCs observed by TEM, there is likely poor loading of PduP1-18-tagged protein into BMCs due to poor formation of intact BMCs in recombinant systems. Our current efforts are focused on improving loading of interior protein into BMCs using alternative microcompartment systems and targeting strategies.
Figure 4. PduP1-18-tagged protein co-isolates with purified bacterial microcompartments (BMCs). (A) Genetic strategy for the PduP1-18-E negative control strain, the PduP1-18-E/Pdu BMC strain, and the PduP1-18-mCherry/Pdu BMC positive control strain from which BMCs were isolated. (B) Gel electrophoresis (SDS-PAGE) and anti-His6 Western blot analysis of purified BMCs. Induction conditions are identified at the top of the gel. Isopropyl-beta-D-thiogalactoside for BMC induction (IPTG); rhamnose for PduP1-18-tagged protein induction (Rha); Pdu BMC shell proteins (red labels). Sizes of molecular weight standards in kilodaltons (kDa) are shown on the left. (C) Transmission electron micrographs (TEM) of purified BMCs. Scale bars, 50 nm.
Purification of Active PduP1-18-E
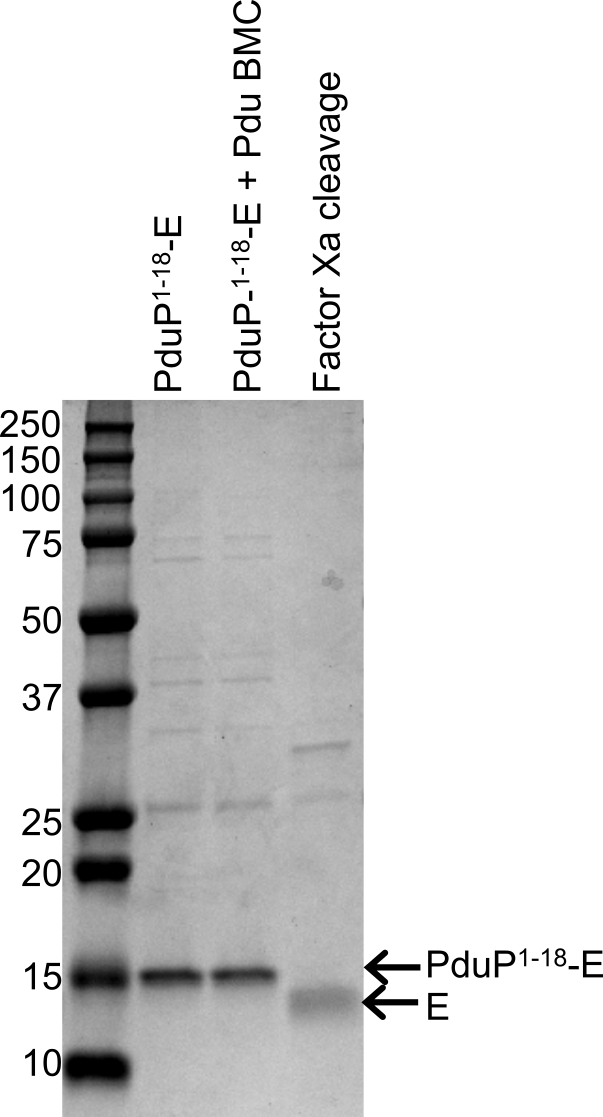
PduP1-18-E was purified to approximately 80 percent purity from cells co-expressing PduP1-18-E and Pdu BMCs (Figure 5). Total yield of PduP1-18-E was 270 µg/L. Following purification of PduP1-18-E, the PduP1-18-tag could be removed from E via Factor Xa proteolysis in a highly efficient manner (88 percent yield). The final yield of non-tagged E was 200 µg/L (approximately 80 percent pure), approximately a seven-fold improvement on a previously reported yield of 27 µg/L (84 percent pure) (Zheng et al. 2009). Purified PduP1-18-E and non-tagged E were both observed to be active in inhibiting Bacillus licheniformis cell growth.
Figure 5. Right side of readout: Purified of PduP1-18-E and non-tagged E. Gel electrophoresis analysis of PduP1-18-E purified from whole cells and non-tagged E after Factor Xa proteolysis. Vertical lanes, labeled left-to-right: PduP1-18-E, purified from cells only expressing PduP1-18-E; PduP1-18-E + BMC, purified from cells co-expressing PduP1-18-E and Pdu BMCs; Factor Xa cleavage, non-tagged E after Factor Xa proteolysis of purified PduP1-18-E. Left side of readout: Sizes (in kilodaltons, kDa) of molecular weight standards.
Impact on Mission
This work demonstrates that re-directed BMC systems can be used to enhance expression of a toxic viral protein, lysis protein E. Our methods can be applied to the purification of other toxic proteins, which are relevant to the biosecurity and biodefense mission goals of the Lawrence Livermore National Laboratory. Broadly, our study has helped to establish engineered BMC systems as a next-generation platform for synthetic biology (Young et al. 2017), which could enable future efforts to develop a novel treatment strategy against antibiotic-resistant bacteria, where bacterial microcompartment encapsulating systems are used to help deliver antimicrobial peptide therapeutics from a genetically-engineered probiotic microbe.
Conclusion
Our research demonstrated the enhanced expression of toxic lysis protein E from bacteriophage phiX174 in Escherichia coli via a re-directed BMC targeting system. Lysis protein E was tagged with various N-terminal BMC targeting sequences, which targeted the protein for association with BMC shell proteins. A PduP1-18-tagged E system appeared to be the best system with the highest expression of the E protein, which was further enhanced by approximately 50 percent by co-expression of Pdu BMC proteins. PduP1-18-E could be optimally expressed at approximately 10 mg/L and purified by Ni-NTA affinity chromatography to 270 µg/L. Non-tagged E was obtained by Factor Xa proteolysis for a final yield of 200 µg of E per liter, almost a seven-fold improvement on values documented in prior reports. Overall, the results demonstrate the utility of re-directed BMC systems to enhance toxic protein yields in recombinant Escherichia coli systems. Our work has contributed to the development of new synthetic-biology capabilities for the Laboratory, as well as future applications towards developing novel countermeasures against biothreat agents.
References
Bernhardt, T. G., et al. 2000. "Genetic Evidence that the Bacteriophage phiX174 Lysis Protein Inhibits Cell Wall Synthesis." Proceedings of the National Academy of Science USA 97 (8):4297―302. doi: 10.1073/pnas.97.8.4297.
Choudhary, S., et al. 2012. "Engineered Protein Nano-Compartments for Targeted Enzyme Localization." PLoS One 7 (3):e33342. doi: 10.1371/journal.pone.0033342.
Chowdhury, C., et al. 2014. "Diverse Bacterial Microcompartment Organelles." Microbiology and Molecular Biology Review 78 (3):438―68. doi: 10.1128/MMBR.00009-14.
Fan, C. G., et al. 2010. "Short N-Terminal Sequences Package Proteins into Bacterial Microcompartments." Proceedings of the National Academy of Science USA 107 (16):7509―7514. doi: 10.1073/pnas.0913199107.
Fan, C. G., et al. 2012. "Interactions Between the Termini of Lumen Enzymes and Shell Proteins Mediate Enzyme Encapsulation into Bacterial Microcompartments."Proceedings of the National Academy of Science USA 109 (37):14995―15000. doi: 10.1073/pnas.1207516109.
Keasling, J. D. 2012. "Synthetic Biology and the Development of Tools for Metabolic Engineering." Metabolic Engineering 14 (3):189―95. doi: 10.1016/j.ymben.2012.01.004.
Lawrence, A. D., et al. 2014. "Solution Structure of a Bacterial Microcompartment Targeting Peptide and its Application in the Construction of an Ethanol Bioreactor." ACS Synthetic Biology 3 (7):454―465. doi: 10.1021/sb4001118.
Parsons, J. B., et al. 2008. "Biochemical and Structural Insights into Bacterial Organelle Form and Biogenesis." Journal of Biological Chemistry 283 (21):14366―14375. doi: 10.1074/jbc.M709214200.
Rosano, G. L., and E. A. Ceccarelli. 2014. "Recombinant Protein Expression in Escherichia coli: Advances and Challenges." Frontiers in Microbiology 5:172. doi: 10.3389/fmicb.2014.00172.
Sargent, F., et al. 2013. "A synthetic system for expression of components of a bacterial microcompartment." Microbiology 159 (Pt 11):2427-36. doi: 10.1099/mic.0.069922-0.
Sinha, S., et al. 2012. "The PduM Protein is a Structural Component of the Microcompartments Involved in Coenzyme B12-dependent 1,2-propanediol Degradation by Salmonella enterica." Journal of Bacteriology 194 (8):1912―1918. doi: 10.1128/Jb.06529-11.
Young, E. J., et al. 2017. "Engineering the Bacterial Microcompartment Domain for Molecular Scaffolding Applications." Frontiers in Microbiology 8:1441. doi: 10.3389/fmicb.2017.01441.
Yung, M. C., et al. 2016. “Methods and Systems to Shield Toxicity During Non-Native Protein Expression and Purification.” U.S. Patent Application 15/178,454, filed June 9, 2016.
Zheng, Y., et al. 2009. "Purification and Functional Characterization of phiX174 Lysis Protein E." Biochemistry 48 (22):4999―5006. doi: 10.1021/bi900469g.
Publications and Presentations
Yung, M. C. 2017. “Re-Directing Bacterial Microcompartment Systems to Improve Expression of Toxic Proteins.” LLNL Biosciences and Biotechnology Division Seminar Series, Livermore, CA, 27 April 2017. LLNL-PRES-729777.
Yung, M. C., et al. 2015. "Engineering bacterial microcompartments (BMCs) to shield toxicity during protein expression and purification." 2015 Synthetic Biology: Engineering, Evolution & Design (SEED) Conference, Boston, MA, 11 June 2015. LLNL-POST-672368.
Yung, M. C., et al. 2016. “Methods and Systems to Shield Toxicity During Non-Native Protein Expression and Purification.” U.S. Patent Application 15/178,454, filed June 9, 2016.
Yung, M. C., et al. 2017. "Re-Directing Bacterial Microcompartment Systems to Enhance Recombinant Expression of Lysis Protein E from Bacteriophage phiX174 in Escherichia coli." Microbiology Cell Fact 16 (1):71. doi: 10.1186/s12934-017-0685-x. LLNL-JRNL-725506.
Yung, M. C., et al. 2017. "Co-Expression of Bacterial Microcompartment (BMC) Shell Proteins Shield Toxicity of Lysis Protein E from Bacteriophage phiX174 During Recombinant Expression in E. coli." 253rd American Chemical Society National Meeting, San Francisco, CA, 4 April 2017. LLNL-POST-728170.