Sergio E. Wong (15-SI-002)
Abstract
The US is a world leader in supercomputing and in harnessing the power of supercomputing for key applications. Our project used supercomputing capabilities to help address a national security problem: pandemic biological threats. The number of pathogens immune to all known treatments is rapidly increasing, and it is increasingly easy to weaponize highly infective bacterial or viral strains. Therefore, the US is constantly vulnerable to virulent strains of known or emerging biological pathogens. During an outbreak, it is imperative to rapidly develop medical countermeasures in the form of an antibiotic or antiviral drug. Unfortunately, swift therapeutic development remains challenging. Our project demonstrated how high-performance computing (HPC) can accelerate development of therapeutic drugs against biological threats. The development of a therapeutic drug is a slow and expensive process. Bringing a drug to market can take more than ten years and cost about $1B. Our work leverages HPC to eliminate a key obstacle for swift development of medical countermeasures: drug induced cardiac toxicity.
In the drug development process, toxicity is a significant problem. If a drug candidate causes toxicity, and in particular potentially lethal toxicity, it will fail and be discarded. Unfortunately, human toxicity does not reveal itself until seven or more years into the drug development process, that is, during human testing. Failure at this stage is disastrous since so much time and money has already been invested; thus prediction of toxicity early in the drug development process is paramount. Drug-induced cardiotoxicity often occurs due to molecular interactions between a drug candidate and a potassium ion channel called hERG. If a drug candidate binds to hERG, a potentially deadly arrhythmia can develop. We built a multi-scale model linking simulations at the molecular level to a clinically relevant outcome, namely an electrocardiogram (ECG) reading. When used as a diagnostic tool, and ECG can capture a particular type of drug-induced cardiotoxicity called long QT syndrome. Long QT syndrome can cause a specific form of ventricular tachycardia, Torsade de Pointes (TdP), which is a potentially deadly condition.
Our integration of models from the molecular scale to a clinically relevant output is the first of its kind. The models span nine orders of magnitude in time and ten orders of magnitude in space. The results of our work can clearly accelerate the development of medical countermeasures to protect the US against human-made or naturally occurring biological threats.
Background and Research Objectives
Within Livermore’s national security mission, the threat of a biological attack justifies the need for medical countermeasures, including targeted therapies. However, the development of novel therapies is riddled with costly failures in clinical trials because of a persistent inability to predict whether a drug will work and whether it will lead to any unintended toxicity. All drugs involve balancing efficacy against toxicity, and clinical trials often reveal toxic side effects so severe that the drug is unusable. Since such failures occur late in the development process, when a drug candidate cannot easily be modified, the trial and the drug fail. Such failures are sufficiently costly and frequent that past failures account for up to 75% of the cost of a new drug.
The heart is one major organ in which toxicity is particularly problematic and difficult to reliably predict. One major cause for cardiotoxicity is molecular interaction between the putative drug and a protein called hERG, which is an ion channel that controls the flow of potassium ions into and out of heart cells. In combination with other ions, potassium flow in and out of cells determines the electrostatic potential that, among other things, helps propagate the electrical signal responsible for heart contraction. Thus, interactions with hERG can affect the electrophysiology and, in some cases, cause arrhythmia. Depending on the severity of the arrhythmia, the consequences can be deadly. Fortunately, ECGs can show warning signs of drug-induced arrhythmia. Specifically, ECGs can show "prolonged QT interval," which is an interval corresponding to the lapse of time between features in an ECG labeled Q and T, respectively.
Our overall objective was to create and utilize a multi-scale, HPC human-heart model to predict the cardiotoxicity of drugs. Without HPC, it would have been impossible to accomplish this objective. The specific goals were as follows:
- Perform simulations of drug-hERG interactions at the molecular scale using atomic-resolution models. The simulations are state-of-the-art and include explicit representations of each atom, water molecule, ion, and membrane structure. The simulation yields data used at the cellular scale.
- Perform simulations at the cellular and tissue level that, based on parameters computed from Objective 1, capture the effects of drug-induced cardiotoxicity.
- Perform whole-organ simulations that use cell and tissue models from Objective 2. Unlike the previous work that only included tissue wedges, our simulations involved an anatomically correct heart. The virtual heart consists of about 300 million cell-like volume elements electronically coupled together. An electrical pulse can propagate through the virtual ventricle and simulate the electrophysiology of a heart beat.
- Simulate a virtual torso that encases the virtual ventricle and computes an ECG-like output. This output is clinically relevant and can be used to predict the toxicity of new drug candidates in healthy and diseased hearts.
Scientific Approach and Accomplishments
Our goal was to develop, validate, and demonstrate an unprecedented, purely computational capability to predict drug-induced cardiotoxicity, using only the drug’s chemical structure to make toxicity predictions. To fulfill this goal, we produced a bottom-up, multi-scale, drug-sensitive heart model. Unlike other approaches, this model is deeply rooted in mechanistic understanding of cardiotoxicity.
Implementation of a multi-scale model to predict drug-induced QT prolongation
The multi-scale model consists of five types of simulations. The first type is the molecular-dynamic simulation, which captures interactions at the molecular level and can be used to produce average parameters necessary for cell level simulations. The second type is the cellular-level simulation. The cell model uses parameters computed from the molecular-level simulations. Several ion channels, such as hERG, are part of the cell model. A dynamic simulation using the cell model yields a time-course of electrostatic potential and current flow from the inside of the cell to the outside. Electrostatic potential and currents produced by one cell trigger the electrostatic potential changes and currents of adjacent cells. The coupling of a group of multiple cells make up a tissue. Tissue level is the next simulation type. At the tissue level, blocks of tissue can be simulated to capture the behavior of sections of cardiac muscle. This leads to the next simulation level: the whole organ. The whole-organ level of simulation shows the propagation of electrical impulses through the anatomically correct tissue. The whole-heart model consists not only of the correct geometry, but also three different cell types which make the model more realistic. To compute an ECG, we simulate a virtual torso around the heart model. It is necessary to simulate a torso because ECGs are measured as differences in electrostatic potential between electrode positions.
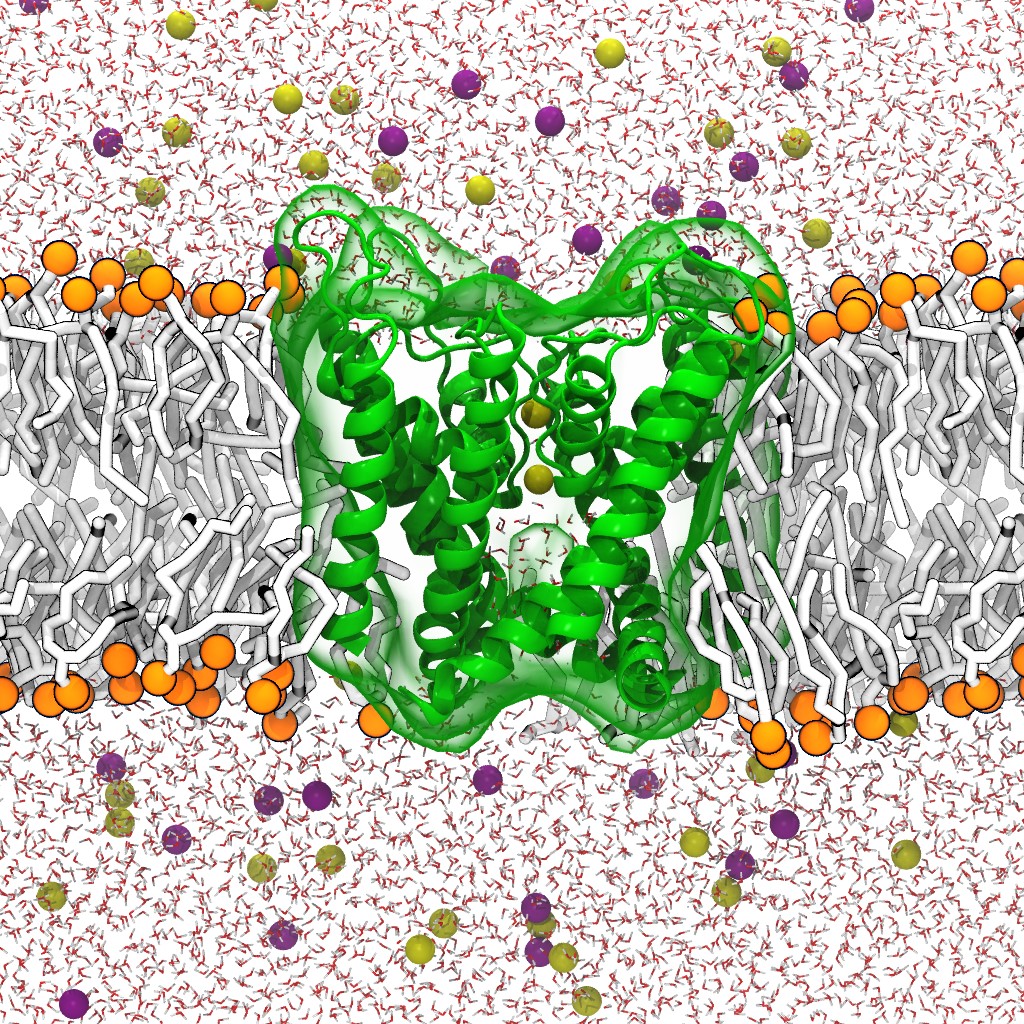
We therefore linked the dynamics of drug interactions at the molecular level to the resulting consequences at the cellular level to model the clinical outcome at the whole-organ level to capture drug-induced QT prolongation. The simulation system is shown in Figure 1. We leveraged the existing Livermore Cardioid model and integrated a more complex cardiac-cellular model and parameters derived from a simulation of molecular interactions to predict the toxic side effect of the anti-arrhythmia drug, ranolazine. Although ranolazine is an antianginal agent showing preference for sodium ion (Na+) channels, ranolazine lacks specificity and also blocks the hERG receptor, causing QT prolongation. We simulated ranolazine binding to the hERG receptor using molecular dynamics and determined binding kinetic rate constants. These simulations informed kinetic parameters for functional scale Markov models of drug interactions with cardiac channels. The perturbed ion channels then affected a cellular model that explicitly included said channels.
Molecular interaction scale
We leveraged HPC resources to implement a robust protocol that uses atomic resolution simulation. The simulation system consists of hERG, ranolazine, and a lipid membrane. We computed hERG-binding kinetic constants using only the molecule’s chemical structure as input. Key to this approach is the atomic-level, physics-based, predictive molecular simulation techniques based on the Markov state model of binding kinetics (Noé and Fischer 2008).
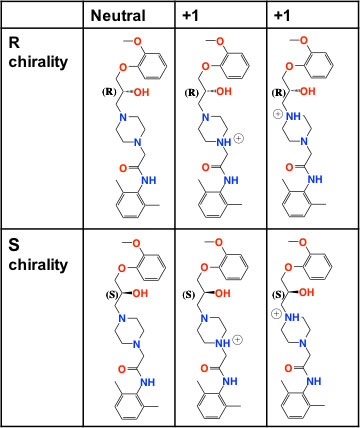
A recent state-of-the art effort utilized approximately 500 independent simulations (50K time steps each) to compute ligand binding kinetics for a single species to an enzyme (Buch, et al. 2011). Binding to hERG, for example, is considerably more complex due to flexibility at the pore entrance. Flexibility can lead to multiple binding configurations and multiple distinct states that need to be modeled. Furthermore, ranolazine is a racemic mixture of two enantiomers (a pair of molecules that are mirror images of each other). Each enantiomer can exist in one of four protonation states, leading to up to eight possible species that must be modeled separately (see Figure 2). Combined with the multiple binding configurations, there could be 12–24 possible binding kinetic parameters to compute.
Figure 2. Distinct chemical species for ranolazine, an anti-arrhythmia drug. We simulated ranolazine binding to the hERG receptor using molecular dynamics and determined binding kinetic-rate constants.
For each bound configuration, stereoisomer, and protonation state, we performed an exploratory atomic resolution simulation explicitly accounting for every atom of the hERG channel, a lipid membrane, and explicit water molecules as solvent. We clustered the resulting trajectories to identify possible sub-states and the resulting clusters were used as starting points for new, independent simulations. The resulting simulations were used for building a Markov state model. The Markov state model consists of sub-states obtained via geometric clustering of the trajectories and transition probabilities or exchange rates among the states.
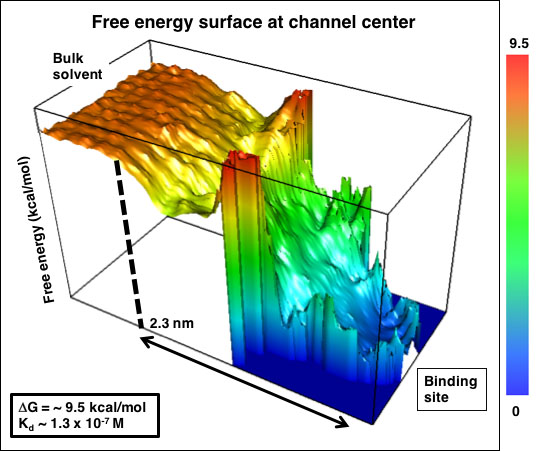
Analysis of the simulations yielded three-dimensional free energy maps (see Figure 3). From these maps, we found the free energy of binding to be 9.5 kcal/mol. It is noteworthy the binding free energy surface flattens at 2.3 nm from the hERG binding site. Electrostatic interactions decrease as the inverse of the distance. hERG transports the positively charged potassium ion. Therefore, it is surprising the overall attraction to the hERG entrance extends only ~1 nm. Using the free energy profile and the diffusion advection equation, we determined the binding-rate constant to be 5 x 108 M-1 min-1.
Figure 3. Energy profile of ranolazine binding to the hERG channel.
Effect of cell-level behavior as a function of drug concentration
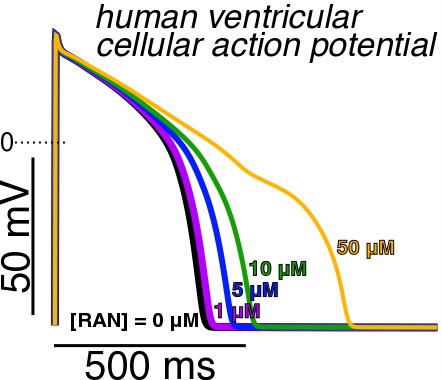
The next step is to link the molecular effects from the molecular-dynamics simulations to the next, larger scale: the cell-level scale. Using the O’Hara-Rudy model, we computed the effect of drug concentration on the cell electrophysiology. The cellular model includes other important ion channels such as sodium, calcium, and other potassium ion channels. The model captures the dynamic, real-time potassium current associated with hERG. Figure 4 shows a definite effect of drug concentration on the behavior at the cellular level.
Figure 4. Electrostatic potential as a function of time for multiple ranolazine concentrations. Ranolazine concentrations include 0, 1, 5, 10 and 50 micromolar.
Tissue and whole organ simulations
The success in the cell-level simulations allowed us to build tissue models. The key step toward tissue and organ simulation was the coupling between millions of cellular models. The tissue model consisted of millions of volume elements associated with individual cell models. Every heart beat involves an electrical pulse that must propagate through the tissue. The electrical pulse diffuses through the tissue according to a diffusion reaction equation. The electrostatic coupling between adjacent volume elements is key. Electrostatic propagation through the tissue model needs to mimic the clinical phenomena. Otherwise, the clinical outcome will be irrelevant.
To perform whole-organ simulations, we used imaging data from the human imaging project. The data was in the form of slices through the heart. Using these slices, we produced a three-dimensional grid where each cell-like volume element was placed. The correct heart anatomy facilitates the correct direction of electrostatic pulse flow. Control whole organ simulations were consistent with known electrophysiology.
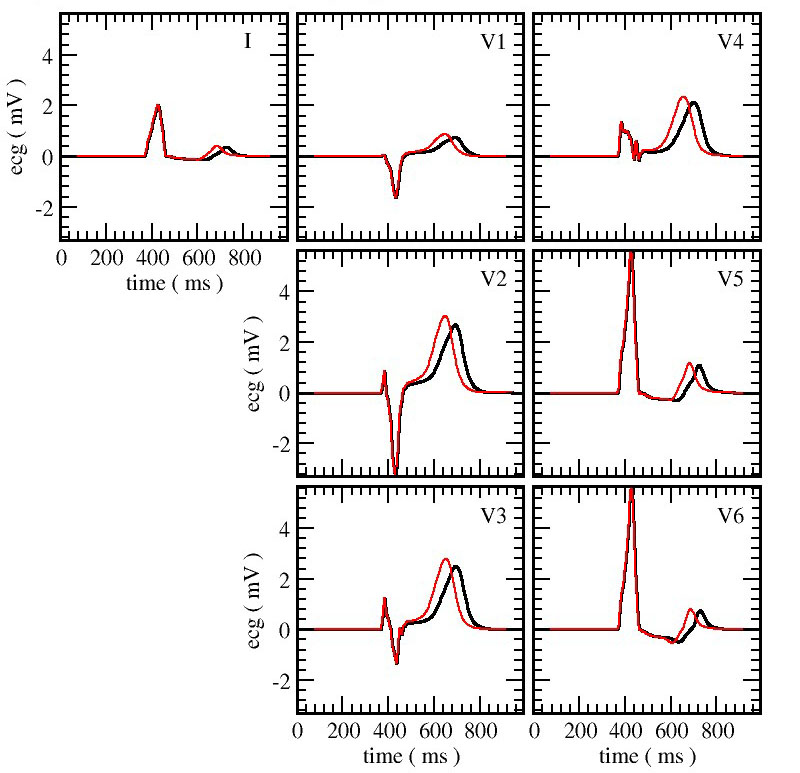
The final step was to compute an ECG-like output. To that end, we used a virtual torso. The virtual heart emits an electrostatic signal that travels through the virtual torso. To make the measurement as realistic as possible, the electrostatic potential differences were measured in the same manner as in the clinic. The results not only show reasonable control calculations, but also the toxic effect of ranolazine (see Figure 5).
Impact on Mission
Our project to develop a virtual human heart that is capable of predicting the pharmacology of drugs will help address the national security challenge regarding pandemic biological threats. The results of our work support Lawrence Livermore's chemical and biological security research and development challenge as well as the NNSA goal of strengthening the science, technology, and engineering base by expanding and applying the nation's science and technology capabilities to deal with broader national security challenges.
Conclusion
During this project, we were able to establish several collaborations and build capabilities to the point where we were able to apply for new funding. We established a collaboration with Joseph Loszcalso, a world-class cardiologist at the Harvard Medical School in Boston, Massachusetts. We are also working with the US Department of Veterans Affairs and have received funding from the American Heart Association.
References
Buch, I., et al. 2011. "Complete Reconstruction of an Enzyme-Inhibitor Binding Process by Molecular Dynamics simulations." Proceedings of the National Academy of Sciences 108 (25):10184–10189. doi:10.1073/pnas.1103547108.
Noé, F., and S. Fischer. 2008. "Transition Networks for Modeling the Kinetics of Conformational Change in Macromolecules." Current Opinion in Structural Biology 18 (2):154–162. doi:10.1016/j.sbi.2008.01.008.
Publications and Presentations
Blake III, R., et al. 2017. “MELODEE: Modular Expression Language for Ordinary Differential Equation Editing." WOLFHPC, Denver, CO, 17 November 2017. LLNL-CONF-737798.