Jeffrey Drocco (16-FS-023)
Abstract
Recently, there has been growing interest in potential uses of gene drives, or mutagenic chain reactions, to effect genetic modifications in wild populations. We computationally demonstrated the feasibility of a novel gene drive design that includes a mechanism capable of later being used to reverse the effect of the drive. Briefly, the “forward” action of the drive proceeds by forcing a genetic change through the population, while the “reverse” action propagates an epigenetic modification that silences the newly introduced genetic elements. These results have implications for the design of controllable gene drive mechanisms that may eventually prove sufficiently safe for uncontained release.
Background and Research Objectives
Geneticists have long discussed the possibility of using “selfish” genetic elements to spread desirable alleles (variant forms of genes) through or eliminate undesirable alleles from a wild population of organisms that are significant from an economic or medical perspective (Burt 2004). Until the discovery of CRISPR/Cas9 genome editing, however, and the fruit fly experiments of Gantz and Bier (Gantz 2015), many proposed mechanisms for driving such an element through a population failed to achieve high efficacy. Since that time, there has been a rapid rise of interest, particularly among insect biologists, in using gene drives for population control of agricultural pests or suppression of human disease vectors (Bull 2017).
Anticipating a need for better tools to understand the impact of an uncontained release of engineered organisms, we conducted this feasibility study as a means of applying Livermore's strengths in computational modeling and simulation to the problem of gene drive propagation. To our knowledge, this project represents the first use of Laboratory supercomputing capabilities for simulation of arthropod population dynamics.
In 2016, the scientific conversation surrounding gene drive technology shifted to the possibility of "reversal" drives (Champer 2016) and more precise mechanisms for controlling the propagation of engineered genetic elements released into a wild population (Noble 2016). In collaboration with scientists from the University of Pennsylvania in Philadelphia, we developed the framework for a new gene drive design that includes the option to introduce an epigenetic modification, which cancels the effect of the drive and spreads at a rate comparable to that of the drive itself. We believe this type of drive design may be useful in producing "biosafe" gene drives whose consequences can be mitigated in the event of unforeseen effects.
Scientific Approach and Accomplishments
Simulation of MCR dynamics
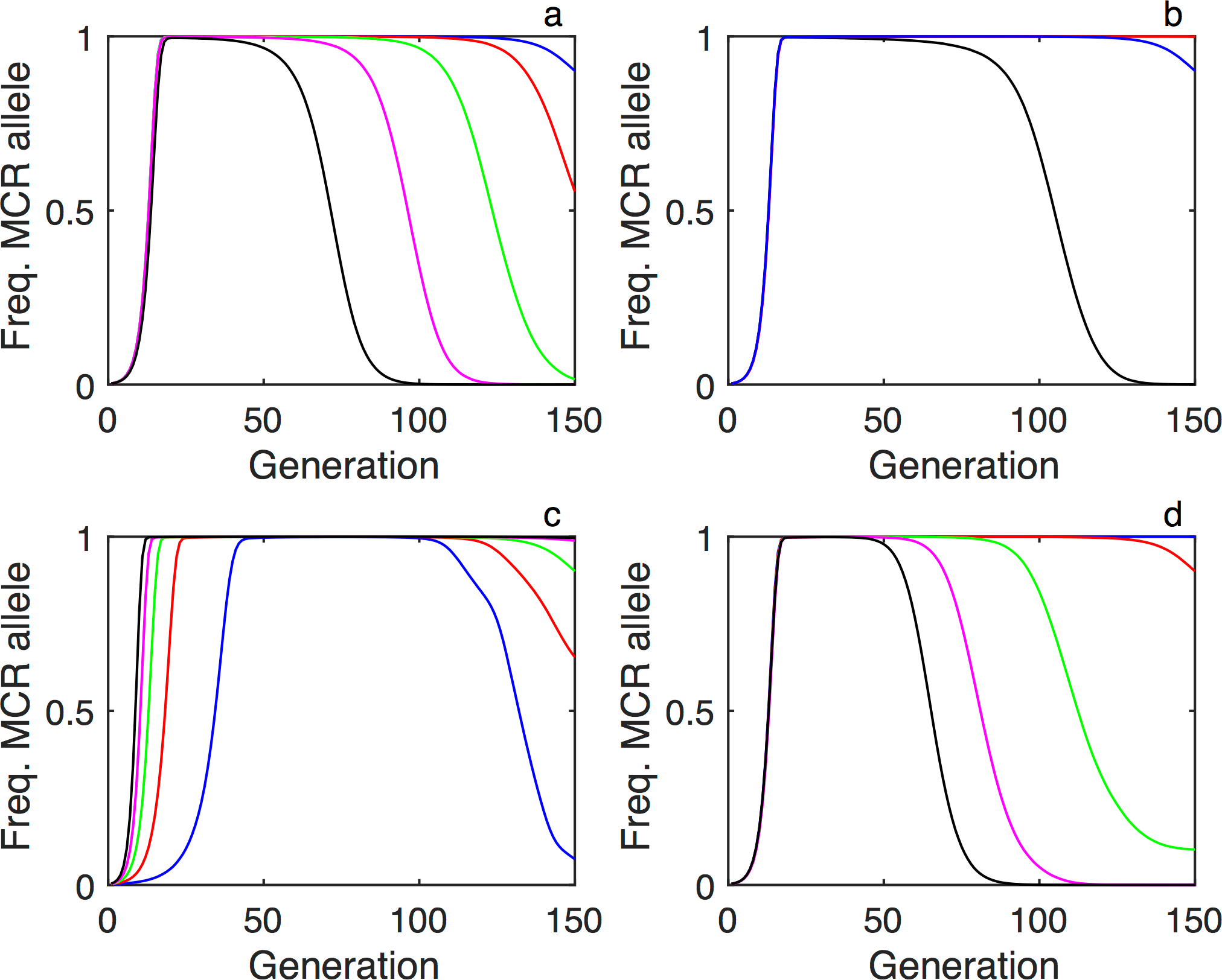
We used a Wright-Fisher model, which captures many of the key features of genetic drift and has played an important role in the development of population genetics, to simulate a population of 50,000 organisms reproducing over 150 generations, in a genetic background that was static excluding the influence of the mutagenic chain reaction (MCR) allele. We identified four primary allelic states: (1) wild-type, (2) the gene drive (MCR) allele, and alleles with resistance to the MCR, produced by erroneous non-homologous end joining (NHEJ) repair of one or more cut sites, that from a fitness standpoint are either (3) costly or (4) cost-free.
In agreement with previous work, we found that drive propagation and stability is enhanced when additional guide ribonucleic acids or gRNAs—short synthetic RNAs composed of a “scaffold” sequence necessary for Cas9-binding and a user-defined ∼20 nucleotide “spacer” or “targeting” sequence which defines the genomic target to be modified—which are targeted to multiple cut sites are included in the MCR cassette (see Figure 1a), when the fitness cost of carrying the MCR allele is lower (see Figure 1c). (A cassette is a type of mobile genetic element that contains a gene and a recombination site. Each cassette usually only contains a single gene and tends to be very small; on the order of 500–1000 base pairs.) Extending previous work, we found that that the stability of the MCR allele in the population is significantly impaired by even relatively small decrements in its fitness relative to the wild-type (see Figure 1d). As expected, we determined that targeting the MCR cassette to genes whose disruption carries a high fitness cost protects against the formation of resistance (Esvelt 2014). More interestingly, however, when the rescue of the essential function of the wild-type by the MCR allele is incomplete, the drive is more stable when the targeted gene is likewise not perfectly recessive lethal (see Figure 1b).
Epigenetic reversal of gene drive effects
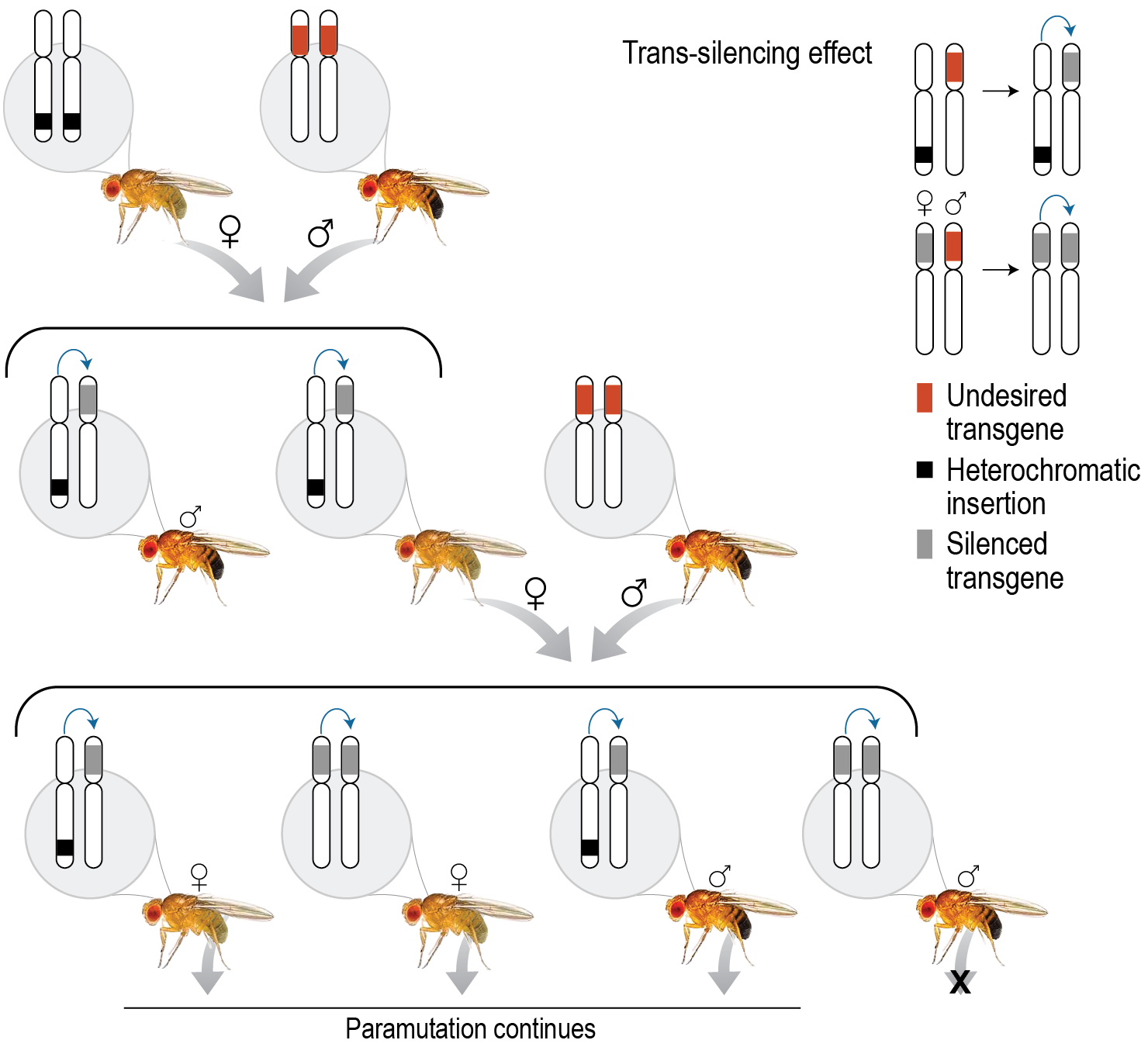
To study mechanisms of gene drive reversal and control, we looked to existing biological pathways that are designed to safeguard the genome from naturally occurring sources of genetic damage. In insects, Piwi-interacting RNAs (piRNAs) play this role by suppressing DNA damage due to the P element (commonly used as mutagenic agents in genetic experiments with Drosophila—a genus of flies) and other invasive mobile DNA elements, called transposons (Khurana 2011). piRNAs are short (24–32 nucleotides), endogenous RNAs that originate from heterochromatic piRNA clusters that silence gene expression by hybridization to transcripts (Ishizu 2012). Importantly, however, scientists have demonstrated that maternal inheritance of piRNAs via the egg cytoplasm can also convert previously dormant transgenic loci into piRNA-producers (Hermant 2015). This silencing phenomenon can propagate exponentially across generations, in an effect called a paramutation (an interaction between two alleles at a single locus, whereby one allele induces a heritable change in the other allele) (see Figure 2). In 2012, scientists demonstrated that this effect could silence the expression of an exogenous transgene for 50 generations (de Vanssay 2012).
We modified our MCR simulation to determine whether paramutations might be used to counteract the effect of a gene drive. Following de Vanssay (2013), we assumed that production of MCR-homologous-piRNAs and suppression of Cas9-mediated gene conversion could be accomplished in the germline by either (1) the presence of a heterochromatic insertion, used to initiate the paramutation process, or (2) the inheritance of MCR-homologous-piRNAs from the maternal cytoplasm.
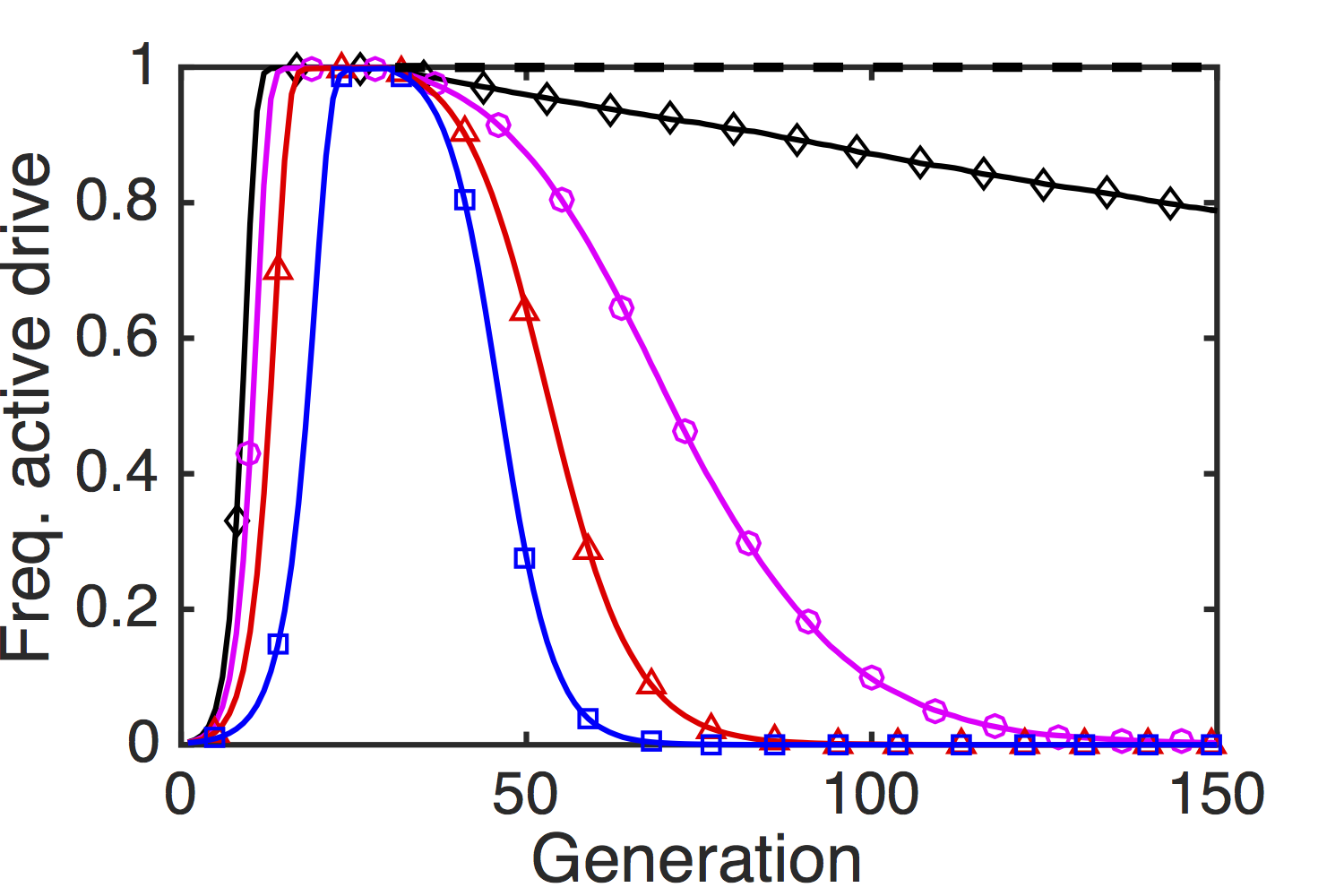
We found that paramutations can rapidly silence the expression of an MCR cassette. Figure 3 shows four gene drive designs, which, despite varying fitness costs associated with the MCR allele, all reach fixation within thirty generations. A piRNA-generating heterochromatic insertion added to a small fraction of the population at that timepoint spread rapidly through the population and effected germline silencing of the drive in all cases where the MCR cassette carried a fitness cost; indeed, the paramutation even spreads, albeit at a slower rate, through the population when the MCR cassette is cost free. This figure indicates the overall level of transcriptional activity associated with the drive; however, if the MCR cassette retains some fitness cost in the silenced state, the genome is also eventually purified of the engineered genetic elements (data not shown).
As even highly efficacious MCR designs can be reversed with the addition of a heterochromatic insertion at a frequency similar to the original, unwanted gene-drive introduction, this may represent an effective strategy for creating novel drives with added protection against undesirable effects in the environment.
Impact on Mission
Our project has contributed to the Lawrence Livermore focus area of chemical and biological security, and builds a novel capability in modeling insect genetics. This work also supported the genesis of a new collaboration with the University of Pennsylvania. Results will enable the Laboratory to better respond to federal sponsor needs as they relate to entomology and novel methods in molecular genetics.
Conclusion
As genetic modification of wild populations become more feasible and cost-effective, computational science has an important role to play in developing basic population dynamics models into a predictive framework that can anticipate both the risks and benefits associated with releases of organisms into the environment. We look forward to deploying Lawrence Livermore supercomputing capabilities to aid experimental efforts currently being germinated with the support of both public and private sector funders.
References
Bull, J. J., et al. 2017. “The Gene Drive Bubble: New Realities.” PLoS Genetics 13 (7):e1006850.
Burt, A., et al. 2004. “Homing Endonuclease Genes: The Rise and Fall and Rise Again of a Selfish Element.” Current Opinion in Genetics and Development 14 (6):609–615.
Champer, J., et al. 2016. “Cheating Evolution: Engineering Gene Drives to Manipulate the Fate of Wild Populations.” PLoS Genetics 17:146–159.
de Vanssay, A., et al. 2012. “Paramutation in Drosophila Linked to Emergence of a piRNA-Producing Locus.” Nature 490 (7418):112–115.
——— 2013. “piRNAs and Epigenetic Conversion in Drosophila.” Fly 7 (4):237–241.
Esvelt, K., et al. 2014. “Emerging Technology: Concerning RNA-Guided Gene Drives for the Alteration of Wild Populations.” eLife 3:e03401.
Gantz, V., et al. 2015. “The Mutagenic Chain Reaction: A Method for Converting Heterozygous to Homozygous Mutations.” Science 348 (6233):442–444.
Hermant, C., et al. 2015. “Paramutation in Drosophila Requires Both Nuclear and Cytoplasmic Actors of the piRNA Pathway and Induces Cis-Spreading of piRNA Production.” Genetics 201 (4):1381–1396.
Ishizu, H., et al. 2012. “Biology of PIWI-interacting RNAs: New Insights into Biogenesis and Function Inside and Outside of Germlines.” Genes and Development 26:2361–2373.
Khurana, J. S., et al. 2011. “Adaptation to P Element Transposon Invasion in Drosophila Melanogaster.” Cell 147 (7):1551–1563.
Noble, C., et al. 2016. “Daisy-Chain Gene Drives for the Alteration of Local Populations.” biorXiv 057307.
Publications and Presentations
Drocco, J. A. 2017. Reversible Gene Drive Mechanism Utilizing Trans-Inactivating Paramutations in Insects. Lawrence Livermore National Laboratory. September 6, 2017. LLNL-CODE-738986.