Gabriela Loots (13-ERD-042)
Abstract
We have developed a new assay method based on accelerator mass spectrometry for correlating genetic variation in animal models of varying bone metabolism with drug response, through the use of calcium-45 and carbon-14 isotope labeling of bone matrix and DNA, respectively. Furthermore, we coupled accelerator mass spectrometry with high-throughput RNA sequencing to determine how human genetics influences metabolism at the cellular level. By treating purified osteoblasts from alleles associated with low-bone-mass phenotypes in humans with several Wnt signal protein ligands known to have bone anabolic responses (Wnt2b, Wnt3a, Wnt4, Wnt5a, Wnt7a, Wnt10b, Wnt11, and Wnt16), we identified several candidates that can now be further investigated for their therapeutic potential (Wnts are glycoproteins that activate intracellular signaling pathways). With our results, we showed that the assay we developed can be used to accurately quantify bone formation and cell proliferation in vivo, to correlate bone metabolic phenotypes with drug response. This assay method can now be further optimized for several other applications including cancer and infectious disease.
Background and Research Objectives
Patients respond differently to the same medication, therefore a great challenge is balancing drug efficacy with toxicity to optimize favorable outcomes of drug treatment. Genetic make-up may account for as much as 90% of this variability in drug disposition and effects. Inter-individual differences in drug response have been shown to be a result of sequence variation in genes encoding drug-metabolizing enzymes, drug transporters, or drug targets. Unlike other factors influencing drug response, inherited determinants remain stable throughout a person's life. Therefore, understanding how the genetic make-up of a person influences drug efficacy and toxicity is key to developing customized effective treatments and minimizing harmful side effects. In addition to influencing drug efficacy, genetics may also contribute to adverse side effects, including death, in up to 5% of the population. Understanding how to predict variation in drug response is a significant gap in our ability to prioritize and optimize new therapeutic entities for development as well as to reduce the costs and number of patients needed for clinical trials. Developing tools to better understand the potential for variation in drug response is an important capability needed for LLNL’s core competency in bioscience and bioengineering and the country’s need to rapidly develop acceptable countermeasures following detection of new chemical, biological, or radiological threats, as well as for new therapies important to public health and the military.
Pharmacogenomics uses genome-wide approaches to understand the interplay between genotype and response to therapeutic treatment. More than 1.4 million single-nucleotide polymorphisms have been identified as part of the human genome project, with over 60,000 affecting genes. Some of these single-nucleotide polymorphisms have already been associated with changes in metabolism or effects of medications and some are now being used to predict clinical response. All drug metabolizing enzymes have genetic variants, many of which translate into functional changes in the proteins encoded. These variants can result in up to 50-fold differences in the rate of drug metabolism among individuals. By identifying pharmacogenetic markers that correlate to a specific genetic make-up, predictions can be made as to how an individual will respond to a particular drug. Because most drug effects are determined by the interplay of several gene products that influence the pharmacokinetics and pharmacodynamics of medications, including inherited differences in drug targets (e.g., receptors) and drug disposition (e.g., metabolizing enzymes and transporters), polygenic determinants of drug effects have become increasingly important in pharmacogenomics, but are also very difficult to collect empirical data or to model. In this project, we aimed to develop novel methodologies for correlating genetic variation in humans with drug response. The findings generated by this research will open new opportunities for designing customized treatments as a function of a person’s genetic makeup with broad applications in biomedical research. Also, this work will generate fundamental knowledge required for new systems biology approaches to studying human tissues in vitro, under different pharmacokinetic conditions, as well as define paradigms that can be applied to future biodefense research.
Scientific Approach and Accomplishments
Identification and Functional Annotation of Wnt Targets in Osteoblasts
Using RNA Sequencing (RNAseq) gene expression analysis, we identified multiple Wnt ligands expressed in bone and selected eight Wnts (Wnt2b, Wnt3a, Wnt4, Wnt5a, Wnt7a, Wnt10b, Wnt11, and Wnt16) for further studies. To identify Wnt targets, we isolated neonatal osteoblasts (bone-forming cells) from the normal bone mass (WT) strain of C57Bl6 calvaria and treated with recombinant Wnt protein for 24 hours and the transcripts quantified by RNAseq. The Wnt-regulated genes were identified by comparing gene expression in Wnt-treated osteoblasts to sham-treated osteoblasts using tuxedo pipeline and voom-limma pipeline.1–3 Using Toppgene software,4 we also performed a gene ontology analysis to determine specific roles played by these Wnts during bone remodeling.
Results
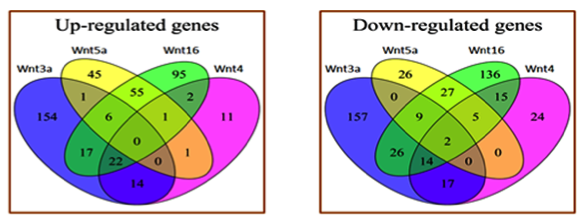
Our analysis identified 879 genes regulated by different Wnts including Wnt3a, Wnt4, Wnt5a, Wnt7a, and Wnt16. Table 1 shows the number of genes up- and down-regulated by each Wnt ligand. Most of the Wnt4 targets overlapped with canonical Wnt3a targets. Target genes regulated by Wnt16 overlapped with both Wnt3a and non-canonical Wnt5a targets. Wnt5a targets had minimal overlap with Wnt3a and Wnt4 targets. These relationships among the different Wnts are shown in Figure 1. Treatment with Wnt7a differentially regulated 72 genes in osteoblasts and most of the Wnt7a targets overlapped with Wnt5a and Wnt16 targets. A gene ontology analysis showed that the Wnts regulate the expression of many genes involved in the regulation of cell proliferation, differentiation, and growth. Wnt5a and Wnt16 also regulated the expression of many cytokines and immune-response-related genes.
| Signal Protein Ligand | Up | Down |
|---|---|---|
| Wnt3a | 214 | 225 |
| Wnt4 | 51 | 77 |
| Wnt5a | 109 | 69 |
| Wnt7a | 45 | 27 |
| Wnt16 | 198 | 234 |
The Role of Wnt Ligand-(Co)receptor Specificity on Target Gene Regulation
Wnt ligands may preferentially bind to certain receptors and coreceptors to regulate different sets of target genes. To determine ligand-(co)receptor-target specificity, osteoblasts isolated from Lrp5KO, Lrp6KO, and Lrp5KO and Lrp6KO mice were treated with the Wnt ligands selected above. The RNA isolated from each Wnt treated sample was sequenced and analyzed as described above to identify genes activated through Lrp5 and Lrp6 and genes that do not require Lrp5/6 for Wnt-induced transcriptional activation.
Results
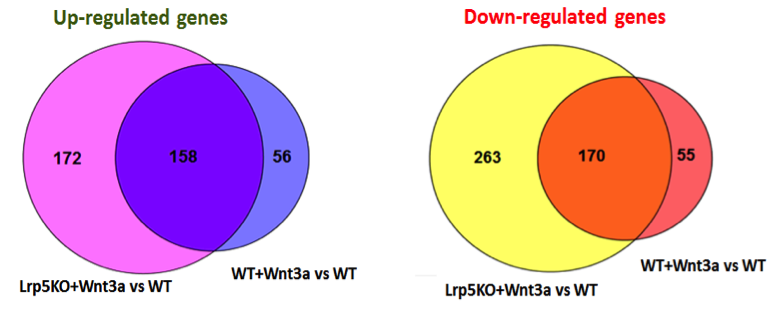
We compared gene expression data from sham-treated Lrp5KO osteoblasts to sham-treated WT osteoblasts and identified 248 genes with altered expression in Lrp5KO osteoblasts compared to WT osteoblasts. Subsequently, we compared gene expression in Lrp5KO osteoblasts treated with Wnts to sham-treated WT osteoblasts and Wnt-treated WT osteoblasts, identifying differentially expressed genes. Figure 2 shows genes shared between the WT osteoblasts + Wnt3a vs WT osteoblasts and Lrp5KO osteoblasts + Wnt3a versus WT osteoblasts. Tuxedo pipeline and voom-limma pipeline together identified 88% of the Wnt3a targets as significantly differentially expressed between Lrp5KO osteoblasts + Wnt3a vs WT osteoblasts. We obtained similar results for genes regulated by Wnt4, Wnt5a, Wnt7a, and Wnt16, suggesting that these Wnts regulate the majority of their target genes independent of Lrp5. It is likely that these Wnts activate most of the target genes through other canonical receptors such as Lrp6 or via the activation of non-canonical pathways.
Calcium-45 Used as a Marker of Bone Formation and Metabolism
Bone metabolism in research models is typically measured by dynamic histomorphometry, a laborious ex vivo approach with limited clinical application. Calcium is a fundamental component of hydroxyapatite, a major component and an essential ingredient of normal bone and teeth, and bone cells that are metabolically active in the balancing of the mineral content of bone. In soft tissues, especially kidneys, calcium levels reflect metabolic processes related to anabolic and catabolic activities of calcified tissues. Calcium tracer kinetic studies can therefore be used to determine relative bone formation and metabolic rates.5 We used the calcium-45 isotope as a tracer to measure bone calcium balance in genetic and chemically induced mouse models of high and low bone mass, to assess differences in bone formation rates via calcium metabolism. Our preliminary data suggests that calcium-45 can be used as a sensitive short-term assay to distinguish mice with different bone mass, because of altered bone formation rates, within 24 hours of a single tracer dose. In addition, calcium-45 tracking can measure bone formation and soft tissue metabolism in response to drug treatment. A single dose of the bone anabolic drug parathyroid hormone6, when used with calcium-45, identified a kidney-dependent phenotype. The successful implementation of calcium-45 assays in rodents could have wide clinical applications because calcium-45 can be safely administered to humans, and potentially could be used as a quantitative assay for risk assessment, early detection, and treatment monitoring of individuals suffering from bone loss. Male mice at 8 to 16 weeks of age were dosed with calcium-45 in a single dose, or a single dose of calcium-45 with a single dose of parathyroid hormone (80 µg/kg dose). High (SostKO, ECR5KO, SostAb-treated), low (Lrp5KO STZ-treated, OPGKO), and WT animals were used. Bone, liver, kidney, blood, and urine were collected and analyzed by liquid scintillation counting. Samples were prepared for counting by complete digestion overnight at RT in 70% nitric acid. Urine and plasma samples were measured without digestion.
Results
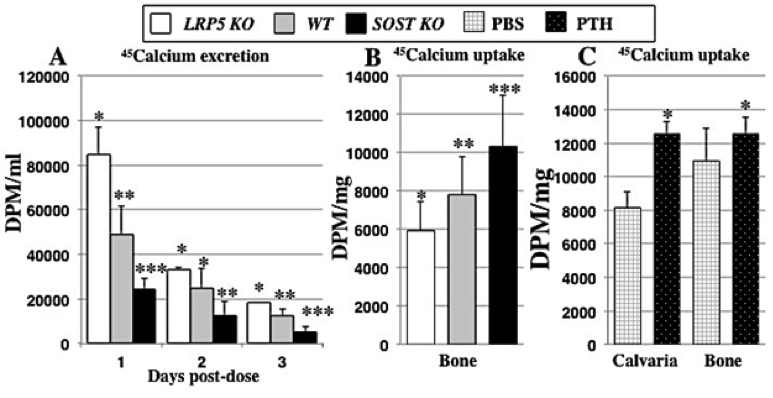
As shown in Figure 3, SostKO mice had over 70% reduced excretion of calcium compared to Lrp5KO mice (p < 0.0001), which is reflective of the high bone-formation rate in SostKO animals and the low bone mass of Lrp5KOmice at peak bone mass (p < 0.01 vs. WT). The reduced calcium-45 in urine of SostKO mice suggested that high bone-formation rates correlate to a significantly higher initial uptake of calcium-45, and conversely, lower excretion. SostKO mice also had significantly higher label retention in bone than Lrp5KO (p < 0.0005) at 24 hours post-dose (p < 0.01 vs. WT) with SostKO having 32% higher calcium-45 bone retention compared to WT. Lrp5KO had 24% lower calcium-45 retention than WT, as shown in Figure 3(A) and (B). Furthermore, we have also validated results for ECR5-/- and SostAb treatment (high bone mass) and OPGKO and STZ type-I diabetic (low bone mass) mice, with each group showing bone retention levels of calcium-45 label 24 hours post-dose that are significantly different from WT (p < 0.05), correlating with the bone formation phenotype of these animals. Based on these results, we next quantified calcium dose response to parathyroid hormone treatment. In bone and urine, we detected significantly different calcium-45 retention and excretion levels in animals treated with parathyroid hormone compared to untreated controls (p < 0.05), as shown in Figure 3(C). Furthermore, the response to parathyroid hormone in bone, kidney, liver, and urine was genotype-dependent, such that SostKO mice showed a different calcium metabolic profile in conjunction with parathyroid hormone treatment than ECR5KO, SostKO, or WT mice (p < 0.01).
Development of Carbon-14–Thymidine for Quantifying Bone Formation and Cellular Proliferation
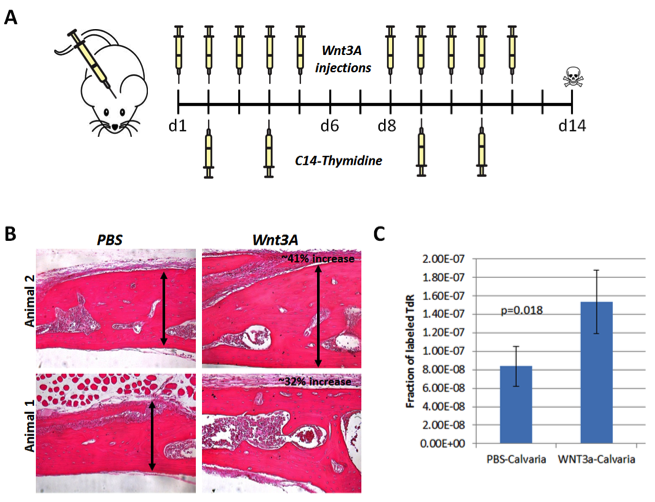
Some Wnt targets may have the potential to promote bone formation. To test candidate secreted molecules identified from the RNAseq analysis above for their ability to enhance bone formation, we first developed a calveria bone formation assay, and used Wnt3A as a proof of principle. Eight-week-old WT mice received Wnt3A injections daily on top of the calvaria on days 1 to 5 and 8 to 12. On days 2, 4, 9, and 11 the mice also received carbon-14–thymidine intravenously, and the calvaria were dissected free of soft tissue on day 14, as shown in Figure 4(A).
Results
Histologically, as shown in Figure 4(B), the calvaria of Wnt3A-treated mice was 32 to 41% larger than the ones in the control group, which were treated with phosphate-buffered saline. When DNA was isolated from these calvaria, and carbon-14 was quantified by accelerated mass spectrometry, we found that Wnt3A-treated samples had a significantly higher radioactive label, indicative of carbon-14 incorporation into DNA at great rate and suggestive of a higher proliferative rate, as shown in Figure 4(C).
Impact on Mission
This project is closely aligned with the Laboratory's national security mission in support of the military and in preparation for a chemical, biological, or radiological terror attack. Lawrence Livermore is a leader in developing new capabilities for biological accelerator mass spectrometry, and this project added the study of bone disease to the portfolio for this spectrometry technology, which is relevant to LLNL's core competency in bioscience and bioengineering.
Conclusion
We have generated "omic" data that can now be used to construct an osteoblast-specific Wnt ligand-(co)receptor-target interaction network, which can be used to pinpoint how specific human alterations in receptor signaling affects signal transduction at the cellular level. This study provided new insights into the Wnt-signaling pathway in the context of bone metabolism and identified new potential pharmaceutical targets for the treatment of osteoporosis, fracture repair, and other bone-thinning disorders. Furthermore, we generated data that suggest that mice can be phenotyped based on their calcium-45 bone metabolic profiles within a 24-hour period, to determine changes in bone formation rates. It is possible to statistically distinguish high bone mass, low bone mass, and normal bone mass from each other, based on uptake, retention, or elimination of calcium-45 from bone. In addition, we have refined these methods to be able to detect bone and soft tissue differences in drug metabolism associated with bone anabolism via parathyroid hormone treatment. We have observed elevated calcium-45 excretion in urine, consistent with a report that documented transient hypercalcemia and hypercalcuria in response to parathyroid hormone treatment, even as we observe the elevated uptake of calcium-45 in bone, consistent with the anabolic properties of parathyroid hormone. We have also been able to link the metabolism of parathyroid hormone to reports of blunted response from parathyroid hormone treatment in SostKO mice. The ability to phenotype both bone metabolism and effects of drug treatment simultaneously with a single assay is a valuable tool for predicting drug responses in different genetic backgrounds, and can uncover phenotypes underlying metabolic differences. In the future, this type of rapid assay will allow screening for unknown bone metabolic disorders, and demonstrates the viability of such methods to screen human patients non-invasively to assess bone metabolism changes from disease or drug treatments.
References
- Trapnell, A., et al., "Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks." Protoc. 7, 562 (2012).
- Ritchie, M. E., et al., "Limma powers differential expression analyses for RNA-sequencing and microarray studies." Nucleic Acids Res. 43(7), e47 (2015).
- Law, C. W., et al., "Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts." Genome Biol. 15(2), R29 (2014).
- Chen, J., et al., "ToppGene Suite for gene list enrichment analysis and candidate gene prioritization." Nucleic Acids Res. 37 (2009). doi: 10.1093/nar/gkp427
- Baron, R., and M. Kneissel, "WNT signaling in bone homeostasis and disease: From human mutations to treatments." Nat. Med. 19,179 (2013).
- Hoeppner, L. H., F. J. Secreto, and J. J. Westendorf, "Wnt signaling as a therapeutic target for bone diseases." Expert. Opin. Ther. Targets 13(4), 485 (2009).