Elizabeth Wheeler | 17-ERD-121
Overview
Cancer is a complex, multi-scale, systemic disease that has continued to elude cure. Currently there is a promising public-private partnership that is taking the approach of using state-of-the-art computational methods and high-performance computing to model cancer. However, the success of these modeling approaches requires the availability of reliable, high-fidelity experimental data to calibrate essential model parameters and validate model predictions. Addressing this challenge, we developed an instrumented human ex vivo three-dimensional tumor model platform that allows for unprecedented spatio-temporal control and measurement of multiple tissue, cellular, and sub-cellular components that drive tumor growth, drug resistance, and metastasis. We characterized the tumor and its microenvironment, including the molecular, cellular, and extracellular matrix and stromal composition. This information was used to recapitulate these tumors in three dimensions using bioprinting to spatially control cellular compartments with integrated multi-scale sensing. Acquired data on response (extracellular and single cell) to a chemotherapeutic drug was leveraged to instantiate an integrated experimental-computational approach to understanding tumor response. This exploratory research and development effort focused on characterizing the tumor and its microenvironment and demonstrating the value of bioprinting tumors.
Background and Research Objectives
During the next decade, over 15 million people (including 150,000 children) will be diagnosed with cancer and more than a third of them will not survive five years past diagnosis with current treatments. For any given chemotherapy, approximately 75 percent of cancer patients will not respond to the treatment (Siegel et al 2018). Furthermore, many of these cancer survivors will develop secondary cancers due to the genotoxic effects of the treatment. A motivating vision for cancer treatment in the future is to use a patient’s own tumor sample, in conjunction with an accurate predictive computational model, to select a personalized and effective therapy. Working towards this goal, and in concert with efforts to bring advanced high-performance computing to cancer therapy, we developed a biomimetic, engineered (instrumented), ex vivo, patient-derived, three-dimensional cancer model. This experimental model maintained a high degree of similarity (spatio-temporal architecture and heterogeneity) to the in vivo human disease and acquired data on intracellular molecular interactions and macro-scale tumor tissue architecture. This data is intended to feed into two ongoing predictive computational efforts: (1) Biological Applications of Advanced Strategic Computing (BAASIC), a multi-institution collaboration studying predictive oncology and (2) Accelerating Therapies for Opportunities in Medicine (ATOM), a collaboration between Lawrence Livermore National Laboratory, Glaxo-Smith Kline, the National Cancer Institute (NCI), and the University of California San Francisco (UCSF) to develop a computation-guided approach to improving preclinical cancer drug discovery and thus identifying the next generation of cancer treatments with better personal outcomes.
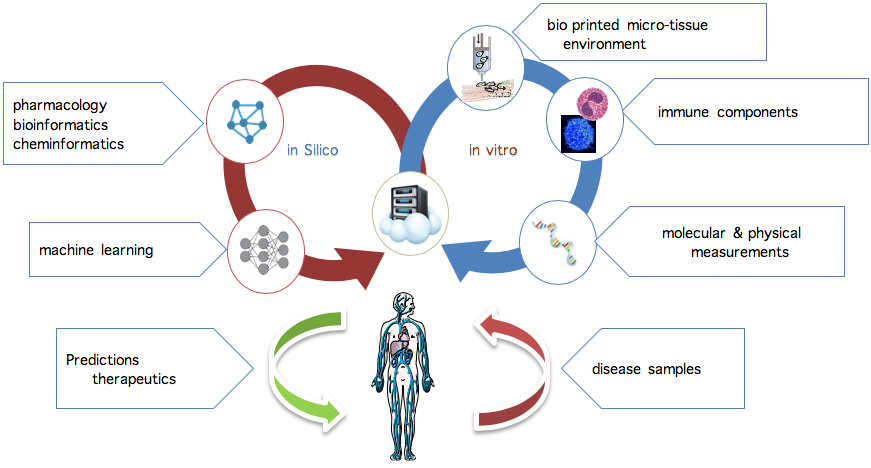
One of the greatest challenges in advancing predictive cancer models and therapeutic screens is the lack of adequate in vitro models that accurately reflect the in vivo tumor microenvironment. Although much has been gleaned from studying two-dimensional human cancer cell lines, these studies offer little predictive value in understanding the complex relationship between tumor composition, microenvironment, and functional outcomes. Growing tumors in three-dimensional (3D) spheroid cultures allows for the study of cell–cell interactions, but these cultures lack reproducibility and true representation of the tumor microenvironment, and they produce limited readouts. A bioprinted ex vivo tumor allows us to physiologically recapitulate all aspects of the tumor environment. The figure below shows the tight iterative approach between computational models in silico and experimental platforms in vitro that is needed to develop a comprehensive predictive biological computation model.
In this project, we leveraged the collaborative strengths and capabilities at Lawrene Livermore and UCSF to demonstrate the viability of the next generation of physiologically relevant 3D tumor models. The next generation of 3D tumor models will ultimately enable the following:
- integration with in-situ and off-line sensor modalities for tissue, cellular, and molecular measurements critical for models
- study of unique configurations to understand the role of tumor microenvironment and extracellular matrix
- investigation of the relationship of the immune system to cancer progression and treatment
- study of tumor evolution
The goal of this work was to demonstrate the feasibility of bioprinting an ex vivo tumor with the characteristic cellular composition of an in vivo tumor and quantifying the differences between the bioprinted and in vivo models. For this proof-of-principle, we focused primarily on mouse cells. Developing protocols to bioprint human cells was beyond the scope of this work.
Impact on Mission
This research advances the science, technology, and engineering competencies that are the foundation of the NNSA mission, as well as the Laboratory's core competency in bioscience and bioengineering. It also addresses the biosecurity mission of the Laboratory in the area of medical countermeasures for rapid mitigation of evolving and unknown threats. Our work specifically supports the institutional initiative on predictive biology through computation and experimentation. Close collaboration with UCSF is also anticipated to seed the growth of new Laboratory–UCSF programs, particularly related to cancer.
Conclusion
We have demonstrated the feasibility of bioprinting tumors and maintaining their viability over the course of several weeks. In addition, we have shown that the bioprinted tumors in the presence of flow are the closest experimental model to the in vivo tumor for the breast cancer cell line explored in this project. Future research will continue to build the complexity of the bioprinted tumor by adding both extracellular support and immune cells to the model. The breadth of the measurements will also be increased to better characterize the physical parameters of both the cells and the environment. This research will ultimately advance our understanding of the role of tumor-immune crosstalk on drug response and resistance by identifying the cellular pathways that determine whether or not tumors grow and the importance of the immune system. By blending innovative technologies with established science and expertise, we are acquiring large, diverse datasets that will aid in modeling tumor growth, evolution, and drug responses, as well as assist patients in making personalized treatment choices. The resultant data are also expected to impact cancer biology by generating prognostic and therapeutic candidate biomarkers predictive of tumor responses to drugs.
References
Siegel, R.L., et al. 2018. "Cancer Statistics 2018." CA: A Cancer Journal for Clinicians 68: 7–30. doi: 10.3322/caac.21442.
Publications and Presentations
Adorno, J. J., et al. 2016. “Tumor On a Chip: Developing a Bioprinted ex-vivo Breast Cancer Tumor Model.” BMES Annual Meeting, Atlanta, Oct. 2018, LLNL-ABS-755436 and LLNL-POST-755439.
Hum, N. R., et al. 2018. “Determining Gene Expression Variability Between in vitro and in vivo Cancer Models: Monolayer, Spheroids, and Mouse Allografts.” AACR, April 2018, LLNL-ABS-74270.
Hynes, W. F., et al. 2018. “Development of a Dynamic Bioprinted Tumor Model.” BMES Annual Meeting, Atlanta, Oct. 2018, LLNL-POST-759650 and LLNL-ABS-750637.