Todd Wasson (15-ERD-053)
Abstract
From electronic medical records to smart cities to computer network traffic monitoring, technological advances have enabled the collection of increasingly massive data sets. The ability to model the systems represented by these data, and especially to make predictions about these systems, can provide immeasurable benefits. In healthcare, for example, accurate and timely prediction of hospitalized patients' future condition would allow better allocation of resources, potentially saving thousands of lives and substantially reducing hospital costs. Within cyber security, effective modeling of network traffic can help identify suspicious behavior and predict and prevent cyber attacks. While the field of big-data analytics has seen substantial advances, it has not kept pace with data-collection advances. Making sense of these data sets and extracting useful information from them remains a challenge. We will develop a framework and a tool set for learning from increasingly prevalent "messy" real-world data, which is often incomplete, heterogeneous, and high-dimensional. Messy data is presently modeled in parts via existing techniques capable of handling a subset of the data, or worse, is discarded outright. We intend to develop models and tools as well as applications for critical data domains. We will develop three independent but complementary tools en route to a unified approach, using cutting-edge cluster computing. The methodological development will be motivated by, and applied to, data sets within healthcare via collaboration with the Kaiser Permanente Medical Groups, as well as to cyber security, with an eye on generalization to other domains.
We expect to develop novel statistical modeling capabilities and tools and ultimately merge these disparate tools and capabilities together to create a cohesive unit. The resulting tools will enable quantifiable prediction based on arbitrary subsets of data, allowing both learning and predictive tasks to fully utilize entire data sets instead of requiring complete observations. Specifically, we will develop the first software tools for (1) modeling time-series data points (a set of observations collected sequentially in time) and nontemporal data together, (2) feature selection on time-series and heterogeneous data together, (3) learning from heterogeneous time-series data with nonrandom missing data, and (4) performing all of these tasks jointly across problem domains. We expect to show significant impact with application to biomedical clinical data, yielding a deployable tool to improve hospitalized patient care. In addition, our software will be relevant to cyber security data, yielding hitherto impossible inference on heterogeneous, potentially unreliably gathered network-sensor data.
Mission Relevance
Harvesting the full potential of important data sets, including cyber and electronic medical record data, and developing a generalized analytic capability that can be broadly applied to other areas of interest to LLNL is aligned with the Laboratory's core competency in high-performance computing, simulation, and data science. The challenges of big data sweep across all mission areas, and immense needs exist to improve capabilities to extract knowledge and insight from large, complex collections of data. Our research also supports the strategic focus area in cyber security, space, and intelligence through predictive analysis of the behavior of complex systems.
FY15 Accomplishments and Results
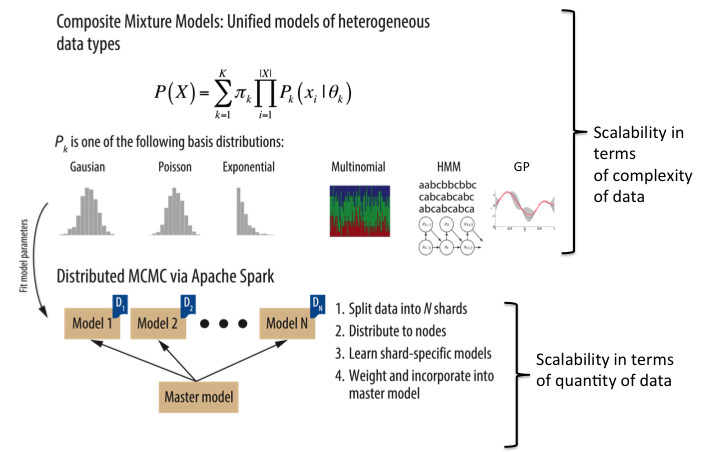
In FY15 we (1) created a Markov-chain Monte Carlo inference approach (algorithms for sampling from probability distributions) for composite mixture models for learning heterogenous data structure and clustering via the Apache Spark cluster computing system; (2) produced two forks of the software to develop time-series inference as well as perform feature selection based on a spike and slab method that is useful for predictions (a type of prior specification used for regression coefficients in linear regression models); (3) worked closely with our Kaiser Permanente Division of Research collaborators to cultivate and procure a customized and rich data set, of which we have begun analysis and application of our tools; and (4) engaged with collaborators from two additional institutions (University of California, San Francisco and University of Virginia) to pursue application of our work to new and differing clinical data sets.
A critical first step to predicting life-threatening sepsis (response to infection) and its outcome is to carefully model patient data, accounting for heterogeneity in the data, and scaling with its size and complexity of the phenomena it describes. We will attempt to both mathematically account for the data complexity and computationally account for its scale by building our approach from the ground up to be compatible with scalable parallel-computing architectures.