Dan Park (16-LW-055)
Project Description
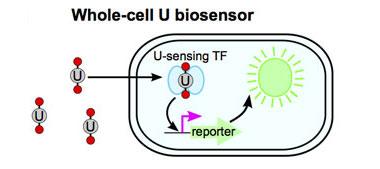
Widespread uranium mining and processing for industrial and military applications over the last several decades has resulted in its significant dissemination in the environment. Uranium is toxic as both a radioisotope and a heavy metal, with exposure leading to nephrotoxicity, genotoxicity, reproductive defects, and diminished bone growth. There is an imperative need to monitor environmental uranium concentrations in a rapid and cost-effective manner to minimize human exposure and to inform remediation strategies. Although current methods such as mass spectroscopy are extremely sensitive and selective, they are costly, low throughput, and cannot be performed on site. Recently, antibodies and deoxyribonucleic-acid (DNA) enzymes with high specificity and selectivity for uranium have been developed, but both systems require sample preparation and costly synthesis or purification steps. In addition, these technologies cannot measure the amount of uranium that is biologically available. We plan to develop whole-cell biosensors that can be used for sensitive, selective, and cost-effective detection of biologically available uranium contamination in the environment. We intend to use synthetic biology to rewire the specificity of native bacterial metal-sensing systems to construct two distinct uranyl ion (UO22+) sensors: cytosolic and periplasmic. Cytosolic and periplasmic refer to the thick solution that fills biological cells and the gel-like matrix in the space between the inner and outer membrane of gram-negative bacteria, respectively. The sensor function will be evaluated in two phylogenetically diverse bacteria and in conjunction with established transducer device technology.
We expect to engineer a novel function in bacterial cells: the ability to directly sense and output a signal in response to UO22+, the most toxic and stable form of uranium in oxygenated environments (see figure). Specifically, we will construct two uranium-sensing regulatory systems that can detect internal UO22+ (cytoplasmic) and external UO22+ (periplasmic). Initially, both biosensors will be built in the rod-shaped, gram-negative bacterium Escherichia coli, a workhorse of biotechnology, and utilize the vast repository of available genetic regulatory parts for this bacterium. This will serve as a proof-of-principle that bacterial regulatory systems can be rewired to produce highly specific and sensitive detection devices for toxins for which there are no known evolved regulatory systems. When coupled with established microelectronics, our whole-cell sensors will provide a simple, quantitative, and cost-effective readout of biologically available uranium contamination in situ in a wide range of environments, facilitating the evaluation of human exposure risks and informing bioremediation strategies.
Mission Relevance
Our work will establish synthetic biology as a powerful tool for chemical detection, thereby expanding the chemical-detection capabilities of LLNL in support of the strategic focus area in chemical and biological security. Our biosensors will provide a significant technical advance towards the rapid and cost-effective monitoring of in situ uranium concentrations, an issue of national-security importance. Lastly, this work will provide an important first step towards the construction of consolidated bioremediators: bacterial systems that possess all the necessary components for deployment in environmental cleanup efforts, relevant to the Laboratory's core competency in bioscience and bioengineering.
FY16 Accomplishments and Results
In FY16 we (1) built an engineered nickel-binding protein system in Escherichia coli, demonstrating effective sensing of its native nickel substrate and using the system to optimize several DNA parts (e.g., nickel-binding protein promoter and reporter promoter); (2) successfully converted the nickel-binding protein system from a repressor into an activator, a significant improvement for sensor deployment; (3) optimized the dynamic range and kinetics of the signal activation process and evaluated uranium-sensing capabilities; (4) identified an optimal output promoter for the PmrAB two-component regulatory system of the bacterium Pseudomonas aeruginosa; (5) swapped the iron-binding domain of the system with three different uranium-binding peptides; and (6) elucidated the genes involved in the regulation of a possible native uranium-sensing system in Caulobacter crescentus, allowing the uranium-sensing performance of this system to be evaluated and further optimized.