Jean-Luc Fattebert (15-ERD-032)
Project Description
First-principles molecular dynamics is an atomistic modeling technique for computing the electronic structure of a molecular system. Including the electrons at the quantum level leads to very realistic models of matter and atomic bonds. However, modeling in materials science with first-principles molecular dynamics is currently limited in atomic size because of its cubic complexity. We are developing, implementing, and validating a truly scalable algorithm for first-principles molecular dynamics simulations of metals, so that one can simulate a number of atoms directly proportional to the number of processors and take full advantage of the millions of processors available on Livermore's BlueGene/Q supercomputer and beyond. We are extending a recently developed algorithm, whose performance grows linearly and in direct proportion to the number of atoms being simulated, to metallic systems by building a general reduced-basis set and by replacing the scalable calculation of selected elements of the inverse of an algebraic Gram matrix with local computation of selected elements of a single-particle density matrix.
Low concentrations of an alloying species can dramatically change the physical properties of alloyed materials, but by definition, modeling low concentrations requires a large total number of atoms and thus, large-size simulations. Furthermore, modeling large defects, such as realistic dislocations produced by radiation damage, requires large atomistic simulations. Currently, realistic modeling of such applications is often out of reach for first-principles molecular dynamics because of their length scale. With our proposed scalable algorithm, very-large-scale molecular dynamics applications will be tractable, which will provide new insights into materials' properties. We are using the new algorithm to compute the single-particle density matrix in the reduced-basis set composed of non-orthogonal localized functions. Exploiting the sparsity of that matrix, we are developing and implementing a truly scalable, first-principles molecular dynamics simulation technology.
Mission Relevance
By developing the next generation of algorithms for first-principles molecular dynamics, we will enable researchers to take full advantage of supercomputers with millions of cores to study physics problems of high interest to Livermore and the DOE—calculating the properties of materials through ab initio modeling. This effort, therefore, supports the Laboratory core competency in high-performance computing, simulation, and data science. This work also addresses the need for better models of materials that can contribute to the Laboratory's work in advanced materials and manufacturing.
FY16 Accomplishments and Results
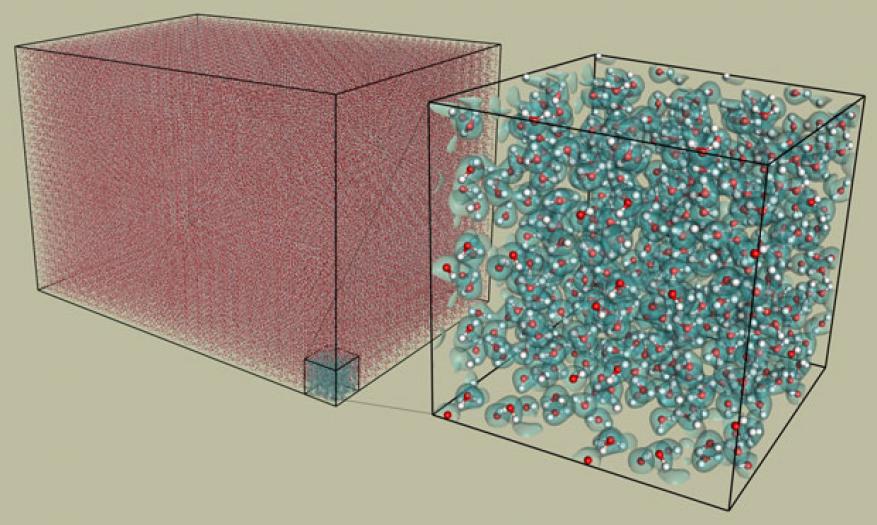
We have implemented various new algorithms to significantly speed up computations, reduce memory, overcome the scaling bottleneck, and enable the very challenging computation of a single-particle density matrix. Specifically, we (1) designed and implemented a new algorithm enabling powers of two in hash-table data access (to allow for complexity), (2) designed and implemented new load balancing for a distributed solver (to increase speed), (3) designed and implemented a new memory-allocation strategy for distributed sparse matrices (to reduce memory), (4) switched parts of the code to single precision (also to reduce memory), (5) implemented and validated new multiple-projector pseudopotentials (to enable computational work with non-valence electrons), and (6) validated, for the first time, the molecular dynamics MGmol code (used to investigate the electronic structure of many-body systems) for the computation of dynamical quantities using a computation of local spatial ordering in liquid water, as illustrated in the figure below. In addition, as finalists for a Gordon Bell award bestowed by the Association of Computing Machinery for an outstanding achievement in high-performance computing, we presented our work at the 2016 International Conference for High Performance Computing, Networking, Storage and Analysis in Salt Lake City, Utah.
Publications and Presentations
- Fattebert, J. L., et al., Modeling dilute solutions using first-principles molecular dynamics: Computing more than a million atoms with over a million cores. Intl. Conf. High Performance Computing, Networking, Storage and Analysis, SC16, Salt Lake City, UT, Nov. 13–18, 2016. LLNL-CONF-689482.