Ali Navid (14-ERD-091)
Abstract
Cells, at all times, need to balance the trade-off between various critical objectives to ensure survival and growth while operating within the constraints of limited resources. Knowing the nature and magnitude of these trade-offs can provide invaluable information regarding the physiological bases behind various observed cellular phenotypes. To gain this knowledge, one needs to conduct system-level analyses of an organism's metabolism. This is primarily achieved via in silico examination of metabolic networks using genome-scale models (GSMs) of metabolism with constraint-based methods like flux balance analysis (FBA).
Unfortunately, FBA always optimizes one biological objective (like growth) as the sole overarching task of a cell. However, studies have shown that biosystems simultaneously optimize multiple objectives in a Pareto optimal (PO) manner. To address this shortcoming of existing methods and realistically model complex biosystems (like microbial communities), we developed an algorithm for conducting high-dimensional Multi-Objective Flux Analysis (MOFA). We have also developed a version of our code that can run on Lawrence Livermore National Laboratory’s high-performance computing (HPC) platforms for cases where very complex MOFA problems require use of parallel computing.
Knowledge about trade-offs among biological objectives also improves our ability to engineer biosystems to produce compounds of interest, such as biofuels or drugs. To highlight this important utility of MOFA, as a test case, we started the process of engineering a synthetic consortium of Clostridium phytofermentans (Cphy), an important biofuel-producing microbe. In the process, we (a) conducted experimental exams to verify catabolite repression in Cphy; (b) developed a GSM for Cphy and used it to identify targets to alleviate catabolic repression; (c) conducted high-dimensional MOFA analyses of Cphy metabolism to identify targets for pathway augmentation in order to improve production of ethanol and hydrogen gas; and (d) designed plasmids for generating single sugar-consuming mutants of Cphy. The plasmids have been transferred to the laboratory of our collaborator where the process of generating the mutants is ongoing.
Background and Research Objectives
The engineering of microbial consortia is a new frontier in synthetic biology. By programming the conduct and performance of select microbial communities we can force these organisms to coordinate their efforts to achieve a specific set of goals, such as the production of compounds of interest like fuels or drugs. Such engineering efforts require system-level understanding of the workings and capabilities of each constituent organism and how they interact in a community. GSMs are one of the key tools used to conduct such systems-biology analyses (Navid 2011; Zomorrodi and Maranas 2012; Stolyar et al. 2007; Henson and Hanly 2014). Although models have been developed for a significant number of single-cell bacteria (e.g., Navid and Almaas 2009; Oh et al. 2007; Feist et al. 2007; Raghunathan et al. 2009; Duarte et al. 2004; Chaudhury et al. 2013), their application for examining metabolic trade-offs within an organism and interactions between individual cells in multi-cellular organisms or microbial communities is limited due to the fact that current constraint-based methodologies like FBA (Orth et al. 2010) examine and optimize only one critical task of the system. In multi-cellular systems, each cell is usually working toward unique goals and the resulting outcome of these efforts could either aid or hinder the goals of neighboring cells. To quantitatively examine the interactions among members of such systems requires use of complex MOFA by means of GSMs.
There have been some attempts at conducting multi-objective optimization (MO) analysis of biological processes using constraint-based models (Handl et al. 2007; Nagrath et al. 2007, 2010; Sendín et al. 2009). However, to date, the maximum number of objective trade-offs that were simultaneously examined have not exceeded five (Nagrath et al. 2007). In some works, in order to study trade-offs among larger groupings of objectives, trade-offs between different combinations of three to five objectives have been examined (Schuetz et al. 2012). While this method works for a small set of objectives, when analyzing larger groups, in order to avoid incomplete considerations of feasible functional capabilities of the system, one would need to analyze an ever-increasing number of small subsets of objectives.
Methods have also been developed, such as ObjFind (Burgard and Maranas 2003), invFBA (Zhao et al. 2016), and BOSS (Gianchandani et al. 2008), that attempt to predict a system’s biological objectives de novo from measured fluxomic data. The solution space for the these methods is highly constrained due to the use of flux measurements. For systems where such information is not available, the solutions provided by these methods would be highly degenerate. For such cases, MOFA has an advantage over these methods because it will provide us with a detailed mapping of the n-dimensional (n=number of examined objectives) Pareto frontier of the objective space. It is also important to note that the solutions provided by the methods discussed above are one of the Pareto solutions provided by MOFA. Thus, for sparsely studied systems, MOFA is the best choice for MO analyses.
The primary goal of this project was to develop an algorithm for automatic generation of MOFA models so that we could analyze trade-offs among diverse biological objectives in biosystems and study the coupled metabolism of diverse organisms in multicellular communities. By the conclusion of the project, we developed three different versions of the code that can be run on different platforms, including one for conducting parallel simulations using Livermore’s high-performance computing (HPC) resources.
We used MOFA to conduct a series of systems biology analyses into biofuel producing capabilities of a number of organisms, including hydrogen gas (H2) producing purple non-sulfur bacterium, Rhodopseudomonas palustris (RP), and lipid-generating algae Chlamydomonas reinhardtii. The results of these analyses have been submitted for publication and have been presented at major scientific conferences.
Additionally, in order to show the advantages of MOFA for purposes of metabolic engineering, we used it to identify target pathways for engineering Cphy. Cphy (Warnick et al. 2002) is an anaerobic bacterium that can directly convert a broad range of biomass sources (e.g., paper sludge, corn stover, grass clippings) into ethanol and H2. One of the most attractive characteristics of this organism is that Cphy can produce biofuels from plant biomass without the need for expensive thermochemical pretreatment and enzyme additions.
Despite all these positive characteristics, we surmised that, like other single cell organisms, Cphy cannot simultaneously consume both hexose and pentose sugars that are released during the process of lignocellulose degradation. This shortcoming (termed catabolite repression) is a significant impediment to the economic use of plant biomass for generation of renewable biofuels (Zaldivar et al. 2001).
We hypothesize that by combining these mutant strains in an engineered consortium (i.e., by compartmentalizing the pathways for consumption of each type of sugar), we will be able to disentangle various hexose and pentose catabolism processes and ensure that only target metabolic pathways consume the limited supply of substrates.
We used FBA and MOFA to identify targets for engineering sugar-specific strains of Cphy. Such detailed analyses of metabolism require use of validated GSMs. To that end, we developed a new, extensively curated GSM of metabolism in Cphy. The model served as the starting point for our MOFA model development and was used for initial selection of gene targets for engineering of substrate-specific organisms.
Finally, once we identified the target genes for metabolic engineering, we designed knockout plasmids for mutant generation using the popular CRISPR-Cas9 genome-editing method (de Souza 2013; Liu and Fan 2014). Simultaneously, we designed plasmids for a backup genome-editing method (group II intron insertion) that previously was used for generating Cphy mutants (Tolonen et al. 2009; Tolonen et al. 2011). We developed the plasmids and introduced them into the Escherichia coli transmission vector. Nonetheless, for both processes we were unable to edit the genome of Cphy. However, our collaborator, Dr. Andrew Tolonen of Genoscope in France, has agreed to attempt to generate the mutants in his laboratory.
Scientific Approach and Accomplishments
1. Development of an algorithm for automatic generation of system-level MOFA models of metabolism for complex biological systems
MOFA is based on the widely used MO method, which is a critical tool in a number of fields where a decision maker needs to consider trade-offs between various conflicting objectives. The outcomes of MO simulations are PO solutions. A PO solution of a problem is one for which any improvement in value of one objective will lead to diminishment of another (Pareto and Bousquet 1964; Pareto 1971).
Our MOFA programs use the Normalized Normal Constraint (NNC) method (Messac et al. 2003) to map the n-dimensional Pareto front of the competing metabolic objectives. The NNC method generates an even distribution of Pareto solutions on both convex and non-convex Pareto frontiers. Additionally, NNC is usable for an arbitrary number of objectives and its results are entirely independent of the scales of the examined objectives.
From the onset, we intended to make our MOFA algorithms user-friendly for general biologists. To this end, we ensured that the required material for doing MOFA using our codes was well described and familiar to computational systems biologists. The required input for our codes are curated FBA models, chemical composition of the growth medium, and biological objectives to be analyzed; the output from the MOFA studies include system-level analysis of metabolic capabilities of microbial community and an ideal metabolic flux pattern for achieving user’s goals.
When combined with FBA, MOFA’s results can be used to identify a set of target genes for metabolic engineering of the system.
Our codes for the first time enable facile high-dimensional MOFA. We have developed the three following versions of the code targeting different user groups:
- A Matlab-based (MathWorks, Inc., Natick, MA) version of the code to be used with the popular COBRA systems biology platform (Schellenberger et al. 2011). COBRA is the primary toolbox used for constraint-based analysis of GSMs. By developing this version of the code, we have made our algorithm available to COBRA’s large user base.
- A C++ version of the code that can use both commercial and open-source linear-program solvers.
- An HPC version of the code that can use Livermore’s parallel-computing resources to simulate MOFA for highly complex problems.
While the first two versions of the code can, in theory, analyze trade-offs among a limitless number of biological objectives (n), time and computational constraints make such simulations (n>15) impractical; however, using our HPC code, we can easily simulate metabolic trade-offs in excess of 50 objectives.
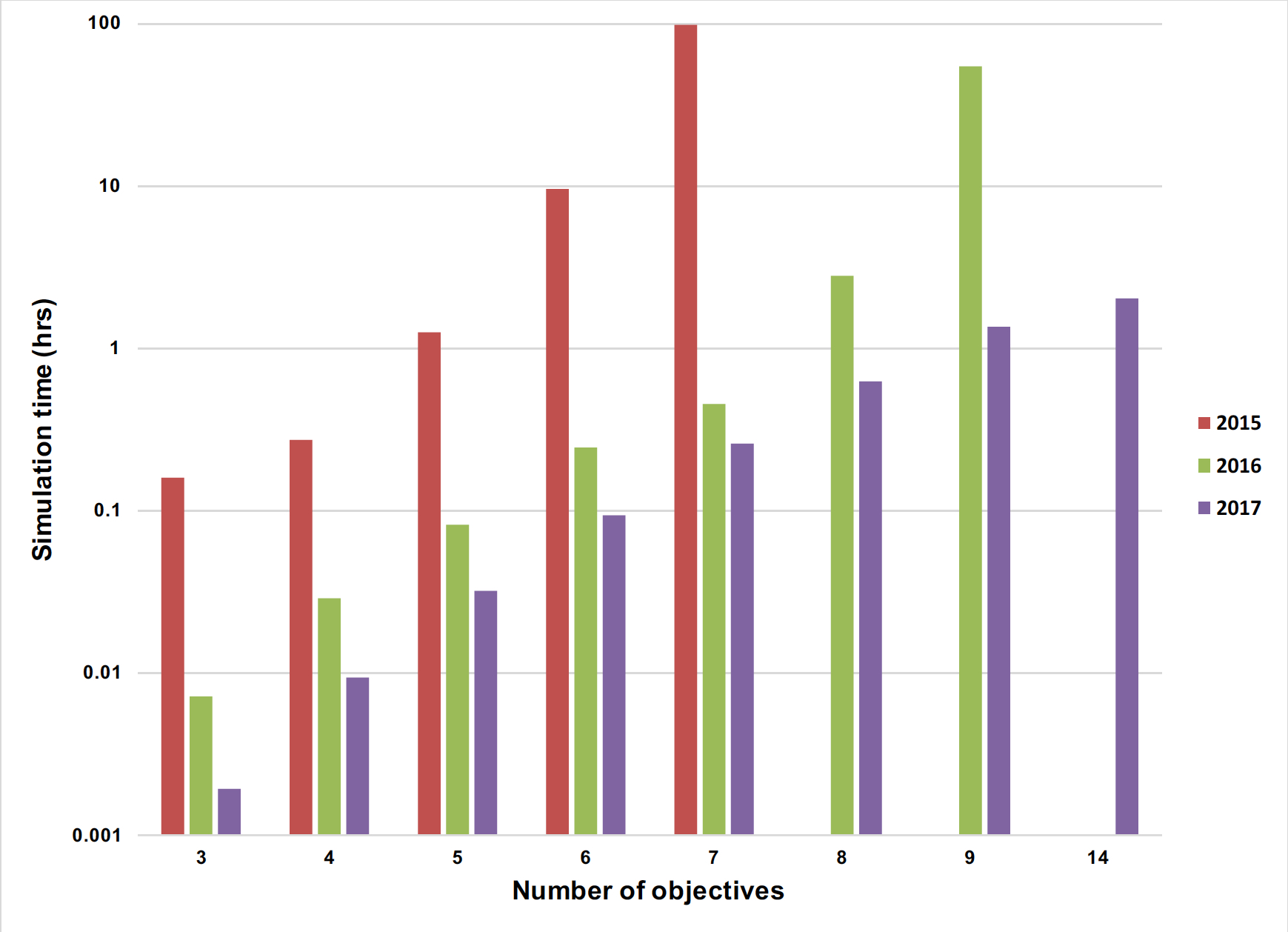
During the past three years we have increased the speed of our non-HPC codes by a couple of orders of magnitude (Figure 1). For comparison purposes, our fastest non-HPC code can simulate a 14-objective MOFA problem in two hours. Meanwhile our HPC code using 4,096 cores can solve the same problem in 18 seconds.
2. Use of tools of computational systems biology to examine the metabolic efficiency and biofuel-producing capability of the model biosystems
During the course of this project, we conducted MOFA examinations of metabolic trade-offs in a number of interesting biofuel-producing organisms. One such organism is the purple non-sulfur bacterium Rhodopseudomonas palustris (RP), a phototrophic organism that is capable of producing H2 as a metabolic byproduct and also metabolizes aromatic compounds as a carbon source in a light-dependent fashion under anaerobic conditions. Examining anaerobic metabolism of RP is environmentally relevant because microbial production of biofuels as well as bioremediation of aromatic pollutants usually occur in low-oxygen environments. We used our code to conduct a high-dimensional MOFA of anaerobic metabolism in RP with the aim of elucidating the trade-offs between the biofuel-producing process and cellular growth. Our results revealed a light-limited growth mode under anaerobic conditions, regardless of the carbon source provided. We found that for photoheterotrophic conditions, RP prioritizes the optimization of carbon efficiency, followed by adenosine triphosphate production and growth rate in a Pareto optimal manner. To achieve maximum carbon fixation, the cell diverts its limited energy resources away from growth and toward fixing CO2, even in the presence of excess reduced carbon. We also found that to achieve the theoretical maximum growth rate, anaerobic metabolism requires import of additional compounds to serve as electron acceptors or sources of protons and oxygen. Finally, we found that H2 production under all circumstances lowers cellular growth rate. The results of these analyses have been submitted for publication. RP is an organism of interest for a joint Livermore-DOE scientific focus area (SFA). We leveraged our MOFA codes, as well as the novel insights we were able to obtain from our analyses of metabolic trade-offs in RP, to establish a new SFA to examine metabolic interactions in various biosystems that are involved in the global carbon cycling.
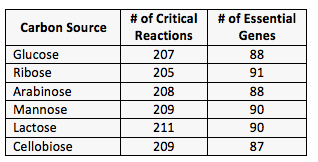
We also used MOFA to study Cphy metabolism. In order to conduct MOFA studies to identify genetic targets for metabolic engineering of sugar-specific Cphy strains, we developed a human-curated model of metabolism in Cphy. We used this GSM with FBA to conduct a series of in silica systems-biology studies of Cphy metabolism. These included assessing the robustness of Cphy metabolism to genetic perturbation while consuming a variety of different sugars (Table 1).
Our results indicate that the robustness of Cphy’s metabolic pathways for consumption of single and complex hexose as well as pentose sugars is nearly identical. This could result from the fact that Cphy’s ecological niche includes both types of sugar as nutrients and hence the organism needs to be adept at using both for survival.
We used our GX-FBA (Navid and Almaas 2012) method to use available gene-expression data to assess changes in metabolism of Cphy as it switches from glucose to other types of sugar (e.g., arabinose). Once again, we found little difference between the metabolism of different sugars.
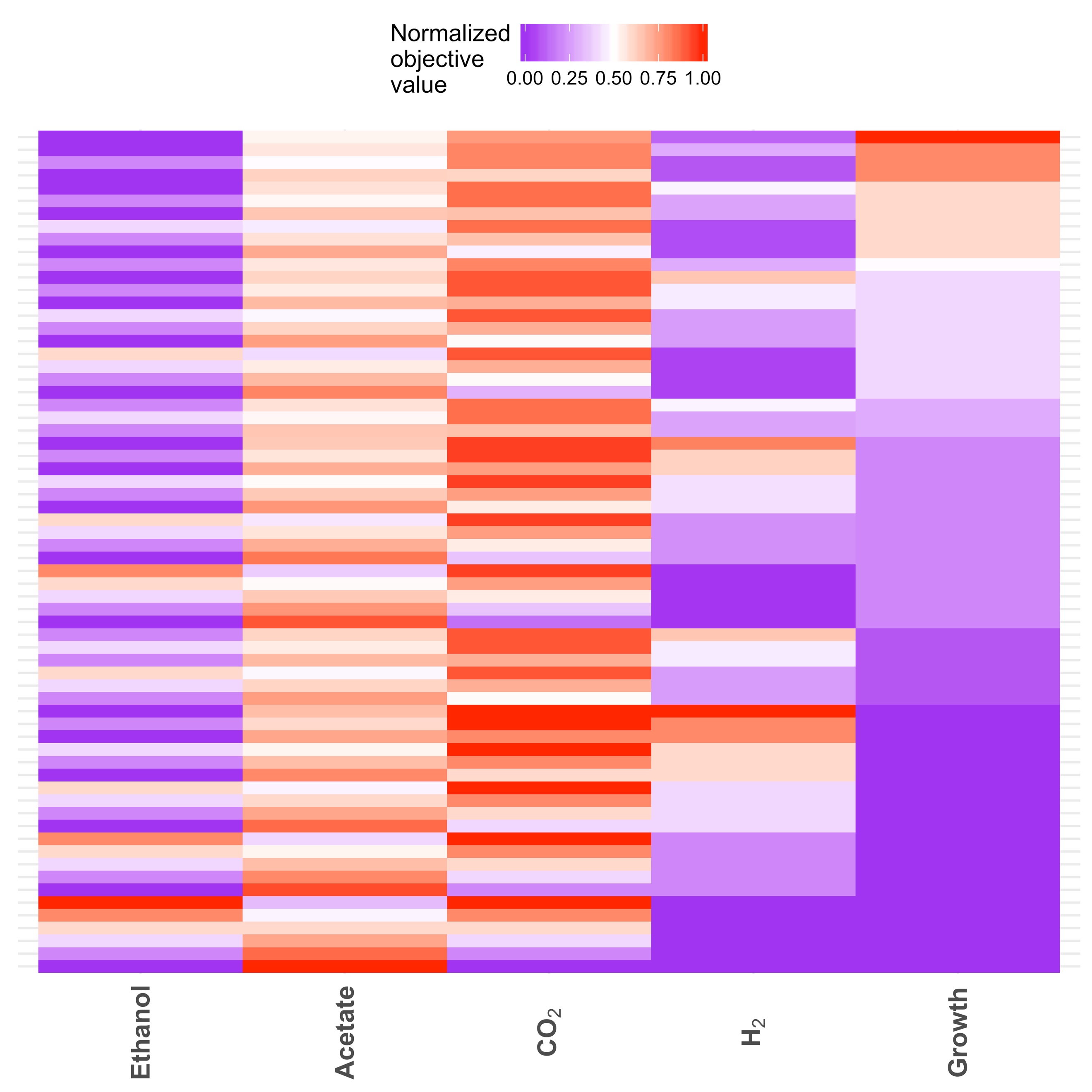
We used MOFA to examine the effects of H2 and ethanol production on growth (biomass yield) of Cphy (Figure 2). Our results show that extra ethanol production is deleterious to cellular growth. However, even at maximum theoretical growth yield, Cphy can produce significant amounts of H2.
3. The metabolic engineering of sugar-specific mutant strains of Cphy
Our first experimental tasks were to (a) verify catabolite repression in Cphy, and (b) measure rates of sugar uptake to constrain our GSM for FBA analyses.
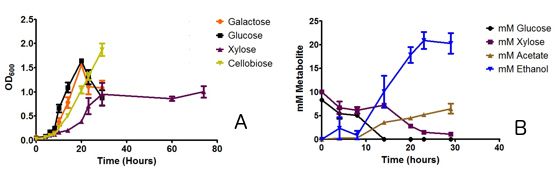
The majority of Cphy liquid cultures were grown on GS2 growth medium (Cavedon et al. 1990) modified for enhanced buffering capacity. Cell numbers were measured by optical density and cell counting. At selected time points, the concentrations of carbohydrates, ethanol, and acetate were quantified using high-performance liquid chromatography (HPLC). A fermentation column (Aminex HPX-87H Column, BioRad) or a HPX-87P column was used for separation of organic acids and carbohydrates. Metabolites were identified by comparison of metabolite-retention times with retention times of authentic standards; metabolite concentrations were calculated using external standard curves generated by measurement of the appropriate standards. Figure 3 shows the result of our analyses. As can be seen, our analyses verified our hypothesis that catabolite repression affects sugar metabolism in Cphy.
To generate the sugar-specific mutants, the first approach we took was the CRISPR-Cas9 genome-editing tool (de Souza 2013; Liu and Fan 2014). Essential elements that make up the primary CRISPR-Cas9 targeting construct include the crRNA and tracrRNA (dual RNA), the donor template, and CRISPR-cas9. Given that our targeted genome Cphy had neither an established mutagenesis protocol nor a CRISPR-Cas9 system, iterative validation of all of the essential elements was necessary to establish the CRISPR-Cas9 system in Cphy (Wang et al. 2015).
The key efforts we made toward implementing CRISPR-Cas9 in CPhy include the following:
- Examined the CPhy genome and determined editing target site(s) within genes of interest. Predicted off-target sites for the sgRNA were checked in both host and E. coli cloning host genome (Naito et al. 2015; Xie et al. 2014).
- Verified the presence of key elements of the cloning vector, including a functioning origin of replication oriT/R, Cas9, a host-compatible promoter upstream of Cas9, a strong RNA promoter upstream of the sgRNA, a homology-directed repair (HDR) cassette, and optimized codon usage.
- Selected an appropriate nuclease Cas9n. The wild type SpCas9 was found toxic to Cyphy, and thus it was desirable to use the Cas9n (HNH, RuvC, or Cas9 D10A) version.
- Selected a strong constitutive promoter for Cas9 that drives the expression of Cas9 in Cphy.
- Selected a promoter to drive the expression of sgRNA. The expression of sgRNA requires a strong sRNA promoter that functions Cphy. Based previously published RNA-seq data, we were able identify a small RNA promoter (sRNA-P) in Cphy to drive the expression sgRNA.
- Designed the HDR template. In order to achieve markerless genome edits, we included the donor DNA in the HDR cassette of the plasmid design. Three CRIPSR-cas vectors were designed and synthesized that specifically target genes of interest in Cphy. Due to a stubborn contamination issue encountered while working in the anaerobic chamber for the selection of antibiotic resistant mutants of Cphy, we were not able to obtain the correct strains consistently, which caused substantial delay in the timeline.
In anticipation of the timing and risks in placing all of our effort on the development of CRISPR in Cphy, toward the end of the first year of the project we started on an alternative gene editing strategy called group II intron retargeting (Tolonen et al. 2009). Based on its intrinsic features, bacterial group II intron target specificity is determined mainly by base-pairing between the target-site DNA and intron RNA. The group II mobility is aided by the presence of an intron-encoded protein (IEP). Expression from pMTL007 or derivative plasmids, of the targeted intron RNA and IEP enables the formation of an RNA-protein complex that inserts the RNA into the targeted DNA sequence and is reverse-transcribed by IEP. This method has been developed for many bacterial species and was recently adapted for use with Clostridium species (Tolonen et al. 2009; Wang et al. 2013; Heap et al. 2007, 2010). Toward this effort, we designed the intron primers necessary for targeting the introduction into genes of interest by submitting the Cphy genome sequence into the algorithm in the intron design site. We also built the PCR fragments into the Intro vector pMTL007. We electroporated the resulting plasmids into Cphy based on a previously published protocol; the resulting mutant was verified by sequencing.
Despite all our efforts, we were unable to generate a sugar-specific mutant strain of Cphy. After conferring with other experts in the field, we were informed that, although some mutant strains of Cphy have been generated, in general Cphy is not an easy organism to genetically modify. One research group that has successfully generated Cphy mutants is that of our collaborator, Dr. Andrew Tolonen of Genescope in France. Even after extensive consultation with Dr. Tolonen’s group, we were unable to generate a mutant. At that point, Dr. Tolonen agreed to attempt to generate the mutants using our plasmids. Currently, that effort is still ongoing. Once the mutants have been developed, in collaboration with Tolonen lab, and using our GSM of Cphy, we will generate the engineered consortium and assess its phenotypic profiles using various carbon sources using other funding.
Impact on Mission
We undertook this task because MOFA simulations are useful for conducting a variety of analyses of immense importance to the Laboratory's critical biosecurity and bioenergy-related missions. These studies can range in scope from examining relationships among organisms that affect global carbon and nitrogen cycles, to factors determining the composition of gut microbiota, and ultimately to host-pathogen interactions.We have already leveraged our code to successfully request funding from DOE to examine multi-species communities that are involved in carbon cycling in nature (both terrestrial and aquatic environments). Also, as a service to the broad scientific community, we aim to offer the MOFA-generating tool as a resource. This will place the Laboratory at the forefront of simulating complex multi-cellular biological systems.
Conclusion
We have developed multiple versions of our MOFA code that can be used on a variety of platforms, including HPC. We used the codes to assess the biofuel-producing capabilities of a number of important organisms, including R. palustris and C. reinhardtii.Although we were not successful in developing the synthetic consortium of Cphy, we (1) verified catabolic repression in Cphy; (2) developed a highly curated GSM for the organism that we used with FBA and MOFA to predict targets for metabolically engineering sugar-specific Cphy strains; (3) designed and acquired plasmids for generating the sugar-specific mutant strains using CRISPR-Cas9 and group II-intron methods; (4) developed collaborations with leading scientists currently studying Cphy metabolism; and (5) ensured that our collaborators are currently using these plasmids to develop the mutants.
We were able to leverage the success of this project to begin further development of the MOFA code. We have presented our MOFA-based results at a number of leading scientific conferences. We have developed a number of successful collaborations. We have submitted two journal articles for publication and two more are being prepared. Based on these results, we believe that this project has made a significant and positive contribution to the Laboratory's mission.
References
Burgard, A. P., and C. D. Maranas. 2003. "Optimization-Based Framework for Inferring and Testing Hypothesized Metabolic Objective Functions." Biotechnology and Bioengineering 82 (6):670―677. doi: 10.1002/bit.10617.
Cavedon, K., et al. 1990. "Cellulase System of a Free-Living, Mesophilic Clostridium (Strain C7)." Journal of Bacteriology 172 (8):4222―4230.
Chaudhury, S., et al. 2013. "Rapid Countermeasure Discovery Against Francisella tularensis Based on a Metabolic Network Reconstruction." PLOS One (8). doi: 10.1371/journal.pone.0063369.
de Souza, N. 2013. "Genetics: CRISPR Silencing." Nature Methods 10 (5):380―381.
Duarte, N. C., et al. 2004. "Reconstruction and Validation of Saccharomyces cerevisiae iND750, a Fully Compartmentalized Genome-Scale Metabolic Model." Genome Resources 14 (7):1298―1309.
Feist, A. M., et al. 2007. "A Genome-Scale Metabolic Reconstruction for Escherichia coli K-12 MG1655 that Accounts for 1260 ORFs and Thermodynamic Information." Molecular Systems Biology 3:121. doi: 10.1038/msb4100155.
Gianchandani, E. P., et al. 2008. "Predicting Biological System Objectives De novo from Internal State Measurements." BMC Bioinformatics 9 (1):43N.
Handl, J., et al. 2007. "Multiobjective Optimization in Bioinformatics and Computational Biology." IEEE/ACM Transactions on Computational Biology and Bioinformatics ACM 4 (2):279. doi: 10.1109/TCBB.2007.070203.
Heap, J. T., et al. 2010. "The ClosTron: Mutagenesis in Clostridium Refined and Streamlined." Journal of Microbiological Methods 80 (1):49―55.
——— 2007. "The ClosTron: A Universal Gene Knock-Out System for the Genus Clostridium." Journal of Microbiological Methods 70 (3):452―464.
Henson, M. A., and T. J. Hanly. 2014. "Dynamic Flux Balance Analysis for synthetic microbial communities." IET Systems Biology 8 (5):214―229.
Liu, L., and X.D. Fan. 2014. "CRISPR–Cas System: A Powerful Tool for Genome Engineering." Plant Molecular Biology 85 (3):209―218.
Messac, A., et al. 2003. "The Normalized Normal Constraint Method for Generating the Pareto Frontier." Structural and Multidisciplinary Optimization 25 (2):86―98.
Nagrath, D., et al. 2007. "Integrated Energy and Flux Balance Based Multiobjective Framework for Large-Scale Metabolic Networks." Annals of Biomedical Engineering 35 (6):863―885.
——— 2010. "Soft Constraints-Based Multiobjective Framework for Flux Balance Analysis." Metabolic Engineering 12 (5):429.
Naito, Y., et al. 2015. "CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced Off-Target Sites." Bioinformatics 31 (7): 1120―1123.
Navid, A. 2011. "Applications of System-Level Models of Metabolism for Analysis of Bacterial Physiology and Identification of New Drug Targets." Briefings in Functional Genomics 10 (6):354―364. doi: 10.1093/bfgp/elr034.
Navid, A., and E. Almaas. 2009. "Genome-Scale Reconstruction of the Metabolic Network in Yersinia pestis, Strain 91001." Molecular Biosystems 5 (4):368―75.
——— 2012. "Genome-level transcription data of Yersinia pestis analyzed with a New metabolic constraint-based approach." BMC Systematic Biology 6 (1):150. doi: 10.1186/1752-0509-6-150.
Oh, Y. K., et al. 2007. "Genome-Scale Reconstruction of Metabolic Network in Bacillus subtitles Based on High-Throughput Phenotyping and Gene Essentiality Data." Journal of Biological Chemistry 282 (39):28791—28799.
Orth, J. D., et al. 2010. "What is Flux Balance Analysis?" National Biotechnology 28 (3):245-8. doi: nbt.1614 [pii] 10.1038/nbt.1614.
Pareto, V. 1971. "Manual of Political Economy." Augustus M Kelley Publishers.
Pareto, V., and G.H. Bousquet. 1964. "Œuvres Complètes: Cours d'économie Politique." Vol. 1: Droz.
Raghunathan, A., et al. 2009. "Constraint-Based Analysis of Metabolic Capacity of Salmonella typhimurium During Host-Pathogen Interaction." BMC Systematic Biology 3:38. doi: 10.1186/1752-0509-3-38.
Schellenberger, J., et al. 2011. "Quantitative Prediction of Cellular Metabolism with Constraint-Based Models: the COBRA Toolbox v2. 0." Nature Protocols 6 (9):1290—1307.
Schuetz, R., et al. 2012. "Multidimensional Optimality of Microbial Metabolism." Science 336 (6081):601-604. doi: 10.1126/science.1216882.
Sendín, J.-O., et al. 2009. "Multi-Objective Optimization of Biological Networks for Prediction of Intracellular Fluxes." 2nd International Workshop on Practical Applications of Computational Biology and Bioinformatics, Spain, 197—205. doi: 10.1007/978-3-540-85861-4_24.
Stolyar, S., et al. 2007. "Metabolic Modeling of a Mutualistic Microbial Community." Molecular Systems Biology 3 (1).
Tolonen, A. C., et al. 2009. "Targeted Gene Inactivation in Clostridium phytofermentans Shows that Cellulose Degradation Requires the Family 9 Hydrolase Cphy3367." Molecular Microbiology 74 (6):1300—1313.
Tolonen, A. C., et al. 2011. "Proteome-Wide Systems Analysis of a Cellulosic Biofuel-Producing Microbe." Molecular Systematic Biology 7.
Wang, Y., et al. 2013. "Development of a Gene Knockout System Using Mobile Group II Introns (Targetron) and Genetic Disruption of Acid Production Pathways in Clostridium beijerinckii." Applied and Environmental Microbiology 79 (19):5853—5863.
——— 2015. "Markerless Chromosomal Gene Deletion in Clostridium beijerinckii Using CRISPR/Cas9 System." Journal of Biotechnology 200:1—5. doi: 10.1016/j.jbiotec.2015.02.005.
Warnick, T. A., et al. 2002. "Clostridium phytofermentans sp. nov., a Cellulolytic Mesophile from Forest Soil." International Journal of Systematic and Evolutionary Microbiology 52 (4):1155—60.
Xie, S., et al. 2014. "sgRNAcas9: A Software Package for Designing CRISPR sgRNA and Evaluating Potential Off-Target Cleavage Sites." PLoS ONE 9 (10):e100448.
Zaldivar, J., et al. 2001. "Fuel Ethanol Production from Lignocellulose: A Challenge for Metabolic Engineering and Process Integration." Applied Microbiology and Biotechnology 56 (1-2):17-34.
Zhao, Q., et al. 2016. "Mapping the Landscape of Metabolic Goals of a Cell." Genome bBology 17 (1):109.
Zomorrodi, A. R., and C. D. Maranas. 2012. "OptCom: A Multi-Level Optimization Framework for the Metabolic Modeling and Analysis of Microbial Communities." PLoS Computational Biology 8 (2):e1002363.
Publications and Presentations
Griesemer, M., and A. Navid. 2016. “HDMOFA: A Tool for High Dimensional Multi-Objective Flux Analysis in Genome-Scale Models of Metabolism." ISMB Conference, Orlando FL, July 8-12. LLNL-ABS-686338.
——— 2017a. “Multi-objective Flux Analysis Software for the COBRA Toolbox”, 253rd ACS National Meeting, San Francisco CA, April 2-6. LLNL-ABS-707597.
——— 2017b. “HDMOFA: A Tool for High Dimensional Multi-Objective Flux Analysis in Genome-Scale Models of Metabolism”, Great Lakes Bioinformatics Conference, Chicago IL, May 15-17. LLNL-ABS-727421.
Navid, A., et al. 2017a. “System-Level Analysis of Metabolic Trade-Offs and Changes During Diurnal Cycle of Chlamydomonas reinhardtii." 2017 Genomic Sciences Program Annual Principal Investigator Meeting, Arlington VA, February 5-8. LLNL-POST-720660.
——— 2017b. “System-Level Multi-Objective Flux Analysis of Metabolism in Clostridium phytoferementans For Optimal Production of Biofuels”, Great Lakes Bioinformatics Conference, Chicago IL, May 15-17. LLNL-ABS-728740.
——— 2017c. “System-level High-Dimensional Multi-Objective Analyses of Metabolic Trade-offs in Biological Systems.”, 25th conference on Intelligent Systems for Molecular Biology and 16th European Conference on Computational Biology, Prague, Czech Republic, July 21-25. LLNL-ABS-729984.