Jonathan Allen (15-ERD-023)
Abstract
Petabytes of genetic data are being collected to document both genetic diversity and the dynamic adaptations organisms make in response to the environment. The current amount of genetic sequence data available for search is 620 billion nucleotides and is estimated to double in size every 18 months. The ability to collect genetic measurement data now outstrips our ability to fully exploit the data to discover new biological insights needed to drive improvements in public health. A key challenge is that searching through billions of bases of eukaryotic genomes remains impossible using conventional tools, and can confound pathogen discovery when differentiating a novel microbe from a novel host genetic variant. We plan to develop new computational methods to store and retrieve functionally significant genetic features. This would be accomplished using a new type of scalable de Bruijn graph, a mathematical concept that turns the formidable challenge of assembling a contiguous genome from billions of short sequencing reads into a tractable computational problem. The new capability would address two problems: identification of features of antibiotic resistance to predict resistance emergence, and detection of host genetic sequences in diverse environments to support pathogen detection. Data structures would be optimized to exploit LLNL's new large-memory high-performance computing environments and will enable linking genetic sequence patterns with experimental outcomes from analysis of much larger genomic data sets than currently possible.
We expect to deliver a new capability to the scientific community that enables searching large genomic data sets against genome databases that were previously considered too large to search. We will design a flexible graph-analytic framework to support scalable analysis of very large genomic data sets, as well as develop bio-surveillance genomic monitoring software and an antibiotic-resistance prediction tool. This will enable identification of organisms in environments that were previously considered impossible to detect and will improve the ability to detect novel pathogens and more accurately characterize infectious disease. In addition, the project would build a new capability for predicting the emergence of antibiotic resistance in clinically relevant environments by tracking the genetic variation associated with a rich set of functional features describing the antibiotic-resistance activities of previously sequenced genes.
Mission Relevance
Our research will address a key need in the Laboratory's bioscience and bioengineering core competency by improving the capability to characterize antimicrobial resistance, anticipate better treatment regimes, and better predict pathogen emergence. The use of novel graph structures and algorithms for genome analysis would be designed to use the unique high-performance-computing resources available at LLNL and advance the state of the art in Livermore's high-performance computing, simulation, and data science core competency.
FY15 Accomplishments and Results
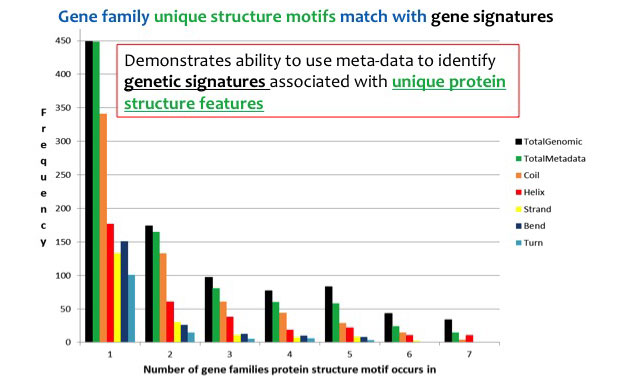
In FY15 we (1) developed a working prototype of the de Bruijn graph and began to demonstrate unique analysis on a scale of thousands of viral genomes; (2) demonstrated the framework using all available Filovirus genomes (including genomes from the recent West African Ebola outbreak); (3) collected, through the use of custom computational pipelines, extensive protein structure meta-data (data that describes other data) for each Filovirus genome to construct a searchable Filovirus genome graph; and (4) began data collection and analysis on antibiotic-resistant genes to be added to a new graph. The graph is designed now to support several different search queries to enable organism and strain identification and function prediction to meet the previously proposed milestones.
Publications and Presentations
- Allen, J., "Using populations of human and microbial genomes for organism detection in metagenomes." Genome Res. 25, 1056 (2015). http://dx.doi.org/10.1101/gr.184879.114